Photo Gallery
Click image to view full version
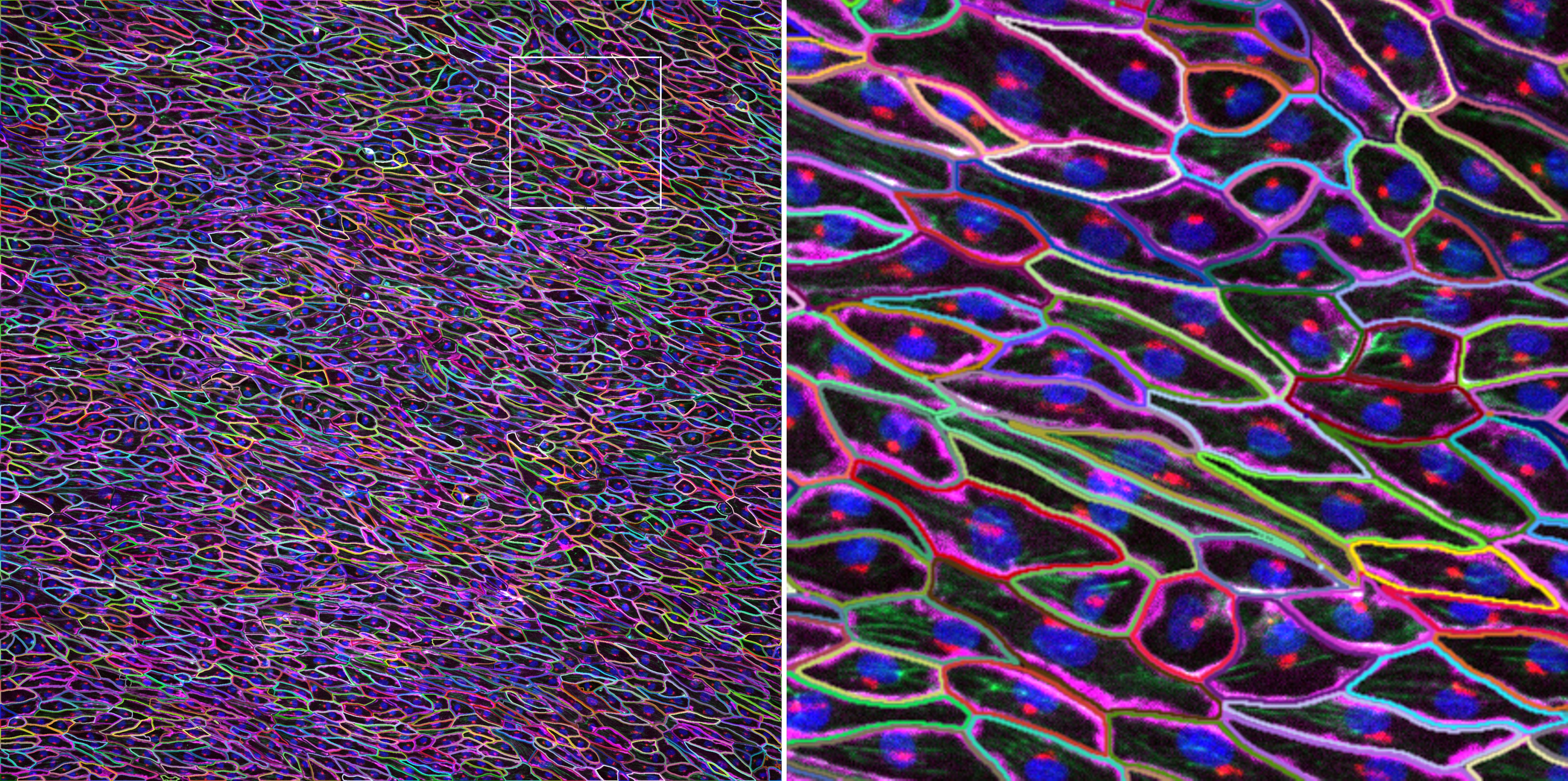
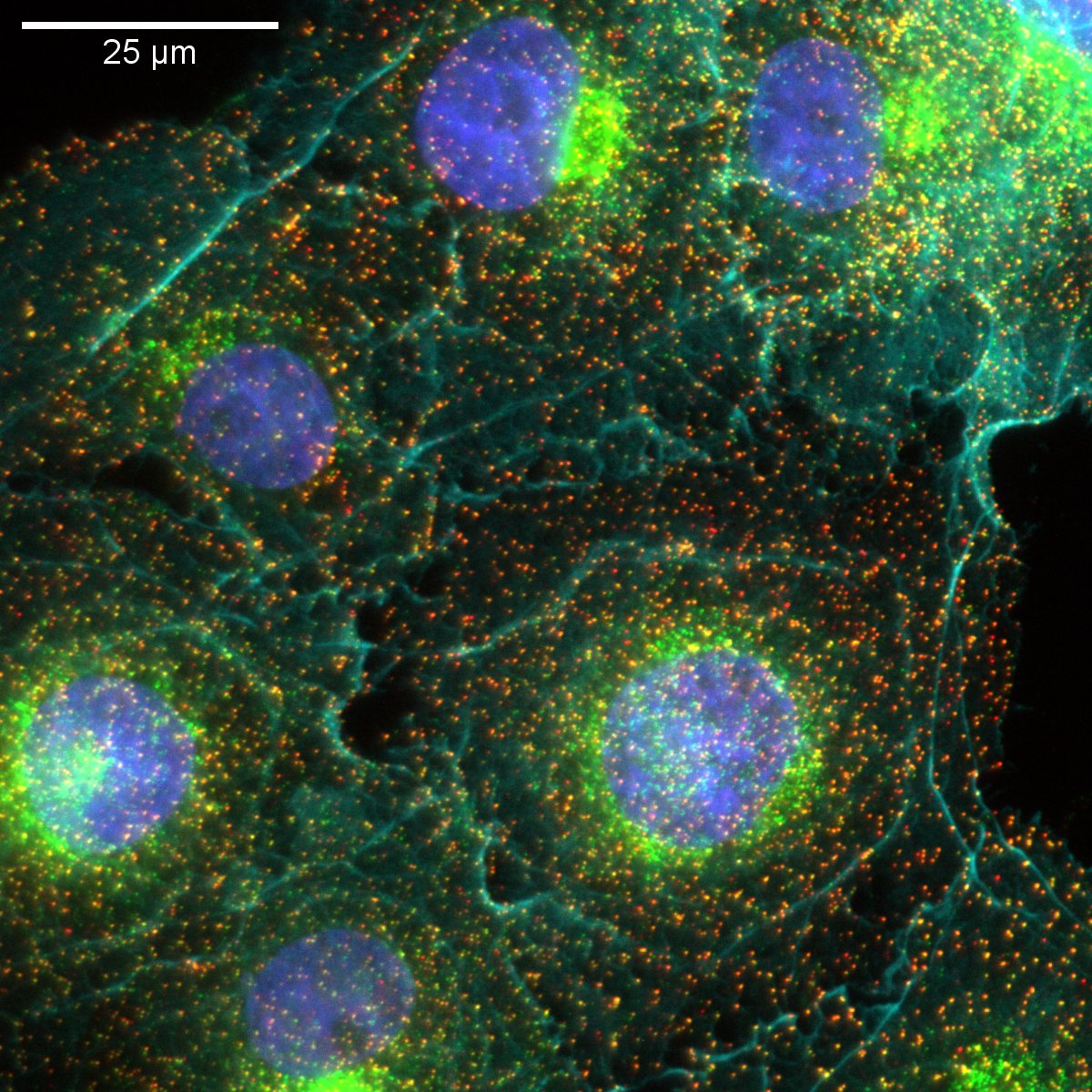
Fiji-based image segmentation
Cells were stained for plasma membrane (CDH5/VE-cadherin, magenta), Golgi (red), actin (green), and nuclei (blue). Imaged on a Nikon CSU-W1 spinning disk confocal microscope using a 10x/0.45 NA objective. Samples were prepared and imaged by Steve Spurgin. Image analysis by Marcel Mettlen.
Cells were stained for plasma membrane (CDH5/VE-cadherin, magenta), Golgi (red), actin (green), and nuclei (blue). Imaged on a Nikon CSU-W1 spinning disk confocal microscope using a 10x/0.45 NA objective. Samples were prepared and imaged by Steve Spurgin. Image analysis by Marcel Mettlen.
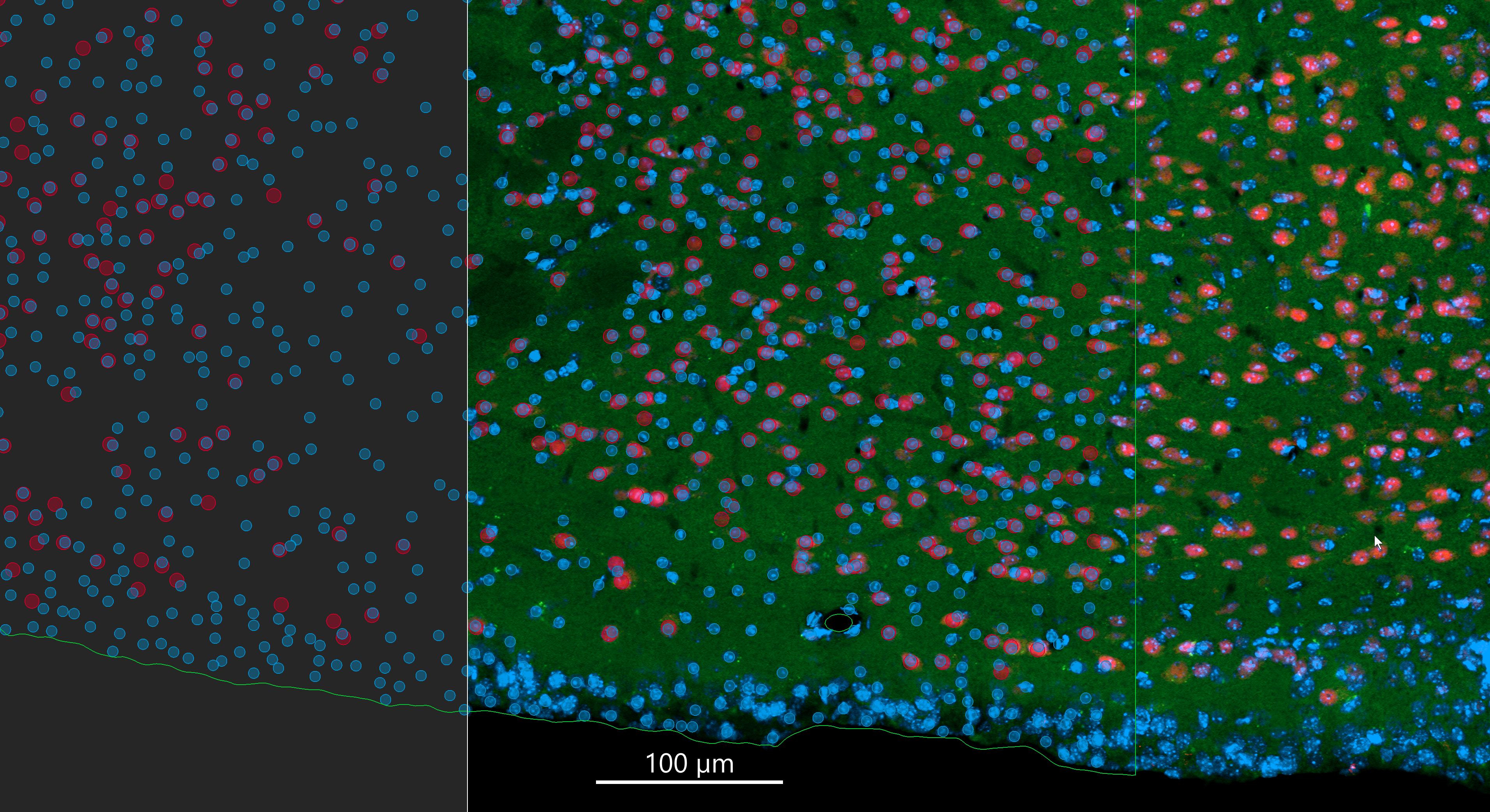
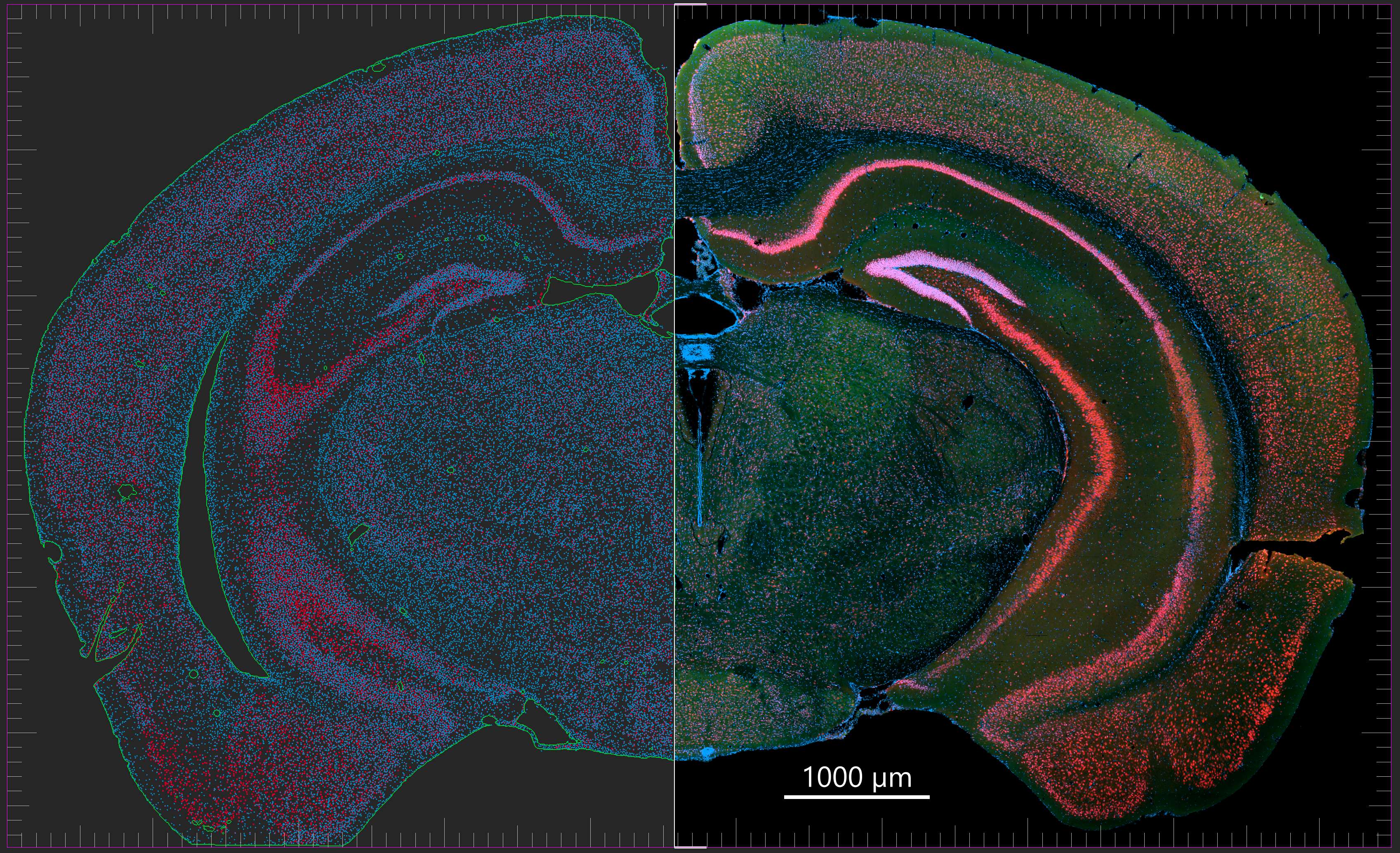
Rendering of frontal mouse brain section
Mouse brain frontal section imaged on a Zeiss LSM 980 with Airyscan 2 and AI Sample Finder. Objective: Plan-Apochromat 25x/0.8 oil immersion. Dataset: 735 stitched tiles; pixel size 0.124 × 0.124 µm; image dimensions 76,822 × 46,842 px; total pixels ~3.6 billion; scan time ~40 min; file size ~27 GB. Image rendered in Imaris 10.2.0. Credit: Chad Smith, Monson Lab.
Mouse brain frontal section imaged on a Zeiss LSM 980 with Airyscan 2 and AI Sample Finder. Objective: Plan-Apochromat 25x/0.8 oil immersion. Dataset: 735 stitched tiles; pixel size 0.124 × 0.124 µm; image dimensions 76,822 × 46,842 px; total pixels ~3.6 billion; scan time ~40 min; file size ~27 GB. Image rendered in Imaris 10.2.0. Credit: Chad Smith, Monson Lab.
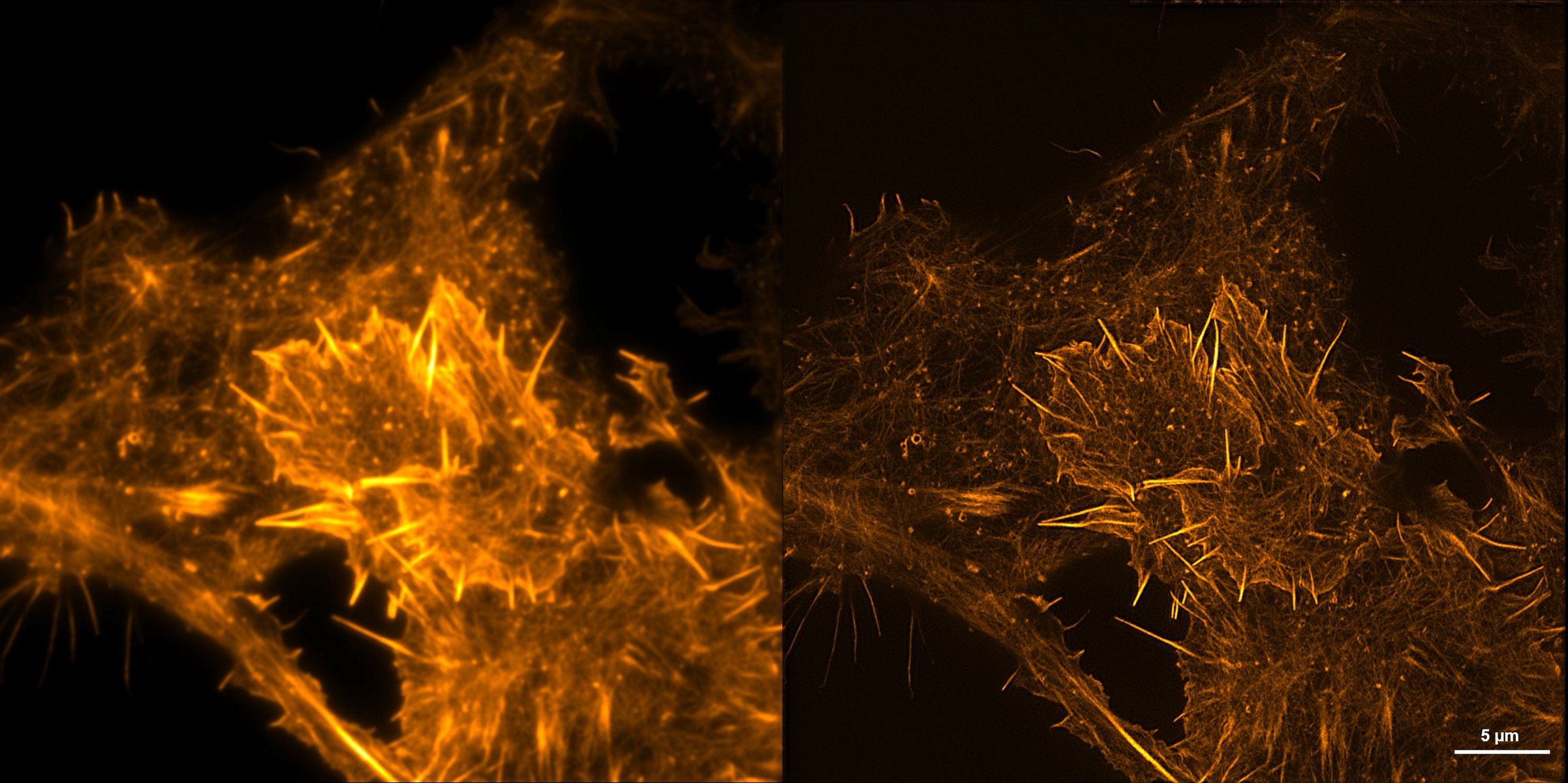
U2OS expressing LifeAct
Live U2OS cells expressing mNeonGreen-LifeAct labeled F-actin. Comparison of TIRF and TIRF-SIM. GE OMX SR; 60x/1.42 NA. Credit: Runwen Yao.
Live U2OS cells expressing mNeonGreen-LifeAct labeled F-actin. Comparison of TIRF and TIRF-SIM. GE OMX SR; 60x/1.42 NA. Credit: Runwen Yao.
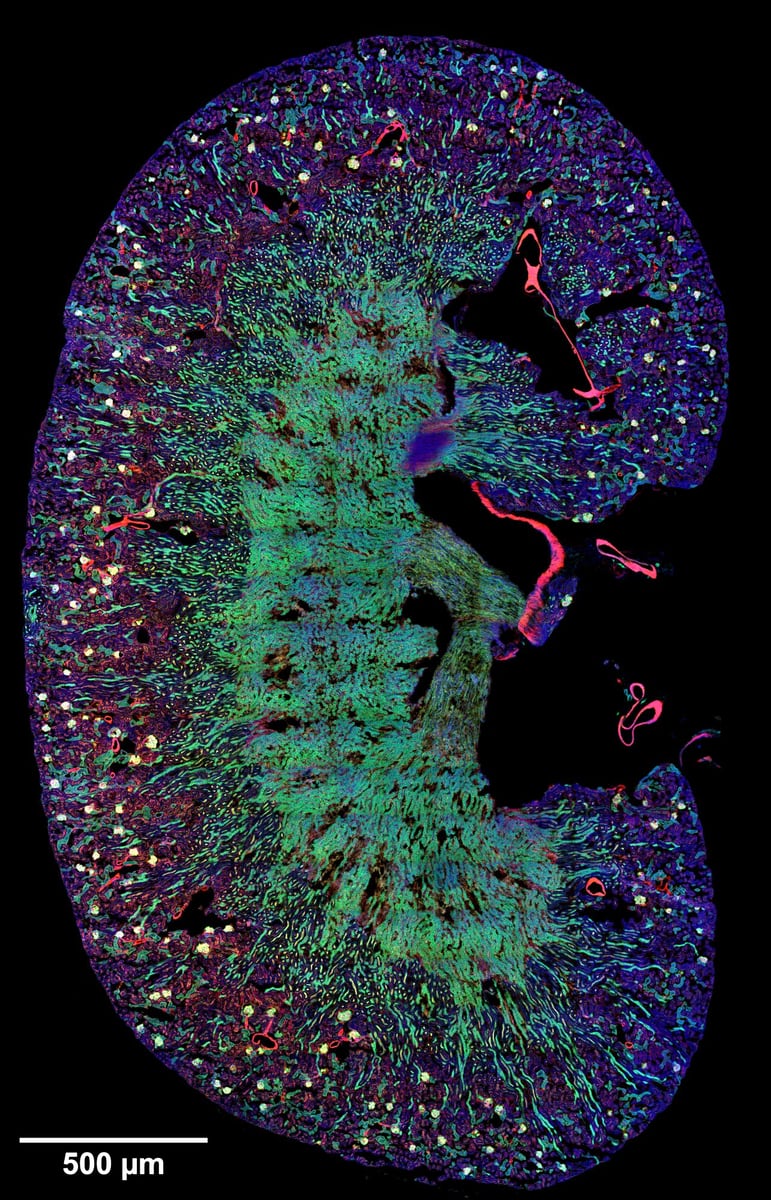
Mouse kidney – overview
Mouse kidney section stained with WGA-Alexa488, Phalloidin-Alexa561, and DAPI. Zeiss LSM 980 Airyscan 2; Plan-Apochromat 20x/0.8 NA; 342 stitched tiles. Credit: Marcel Mettlen.
Mouse kidney section stained with WGA-Alexa488, Phalloidin-Alexa561, and DAPI. Zeiss LSM 980 Airyscan 2; Plan-Apochromat 20x/0.8 NA; 342 stitched tiles. Credit: Marcel Mettlen.
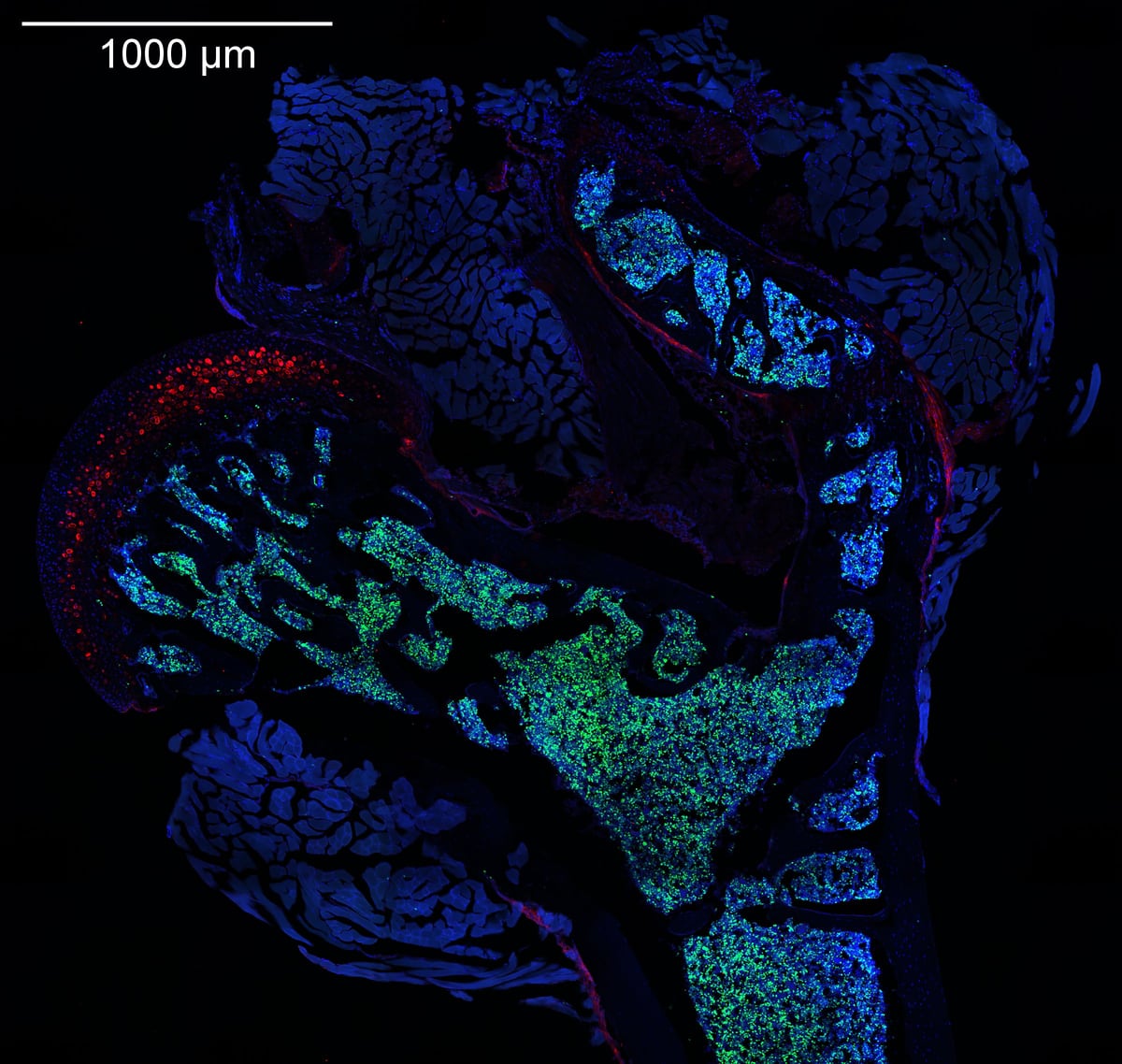
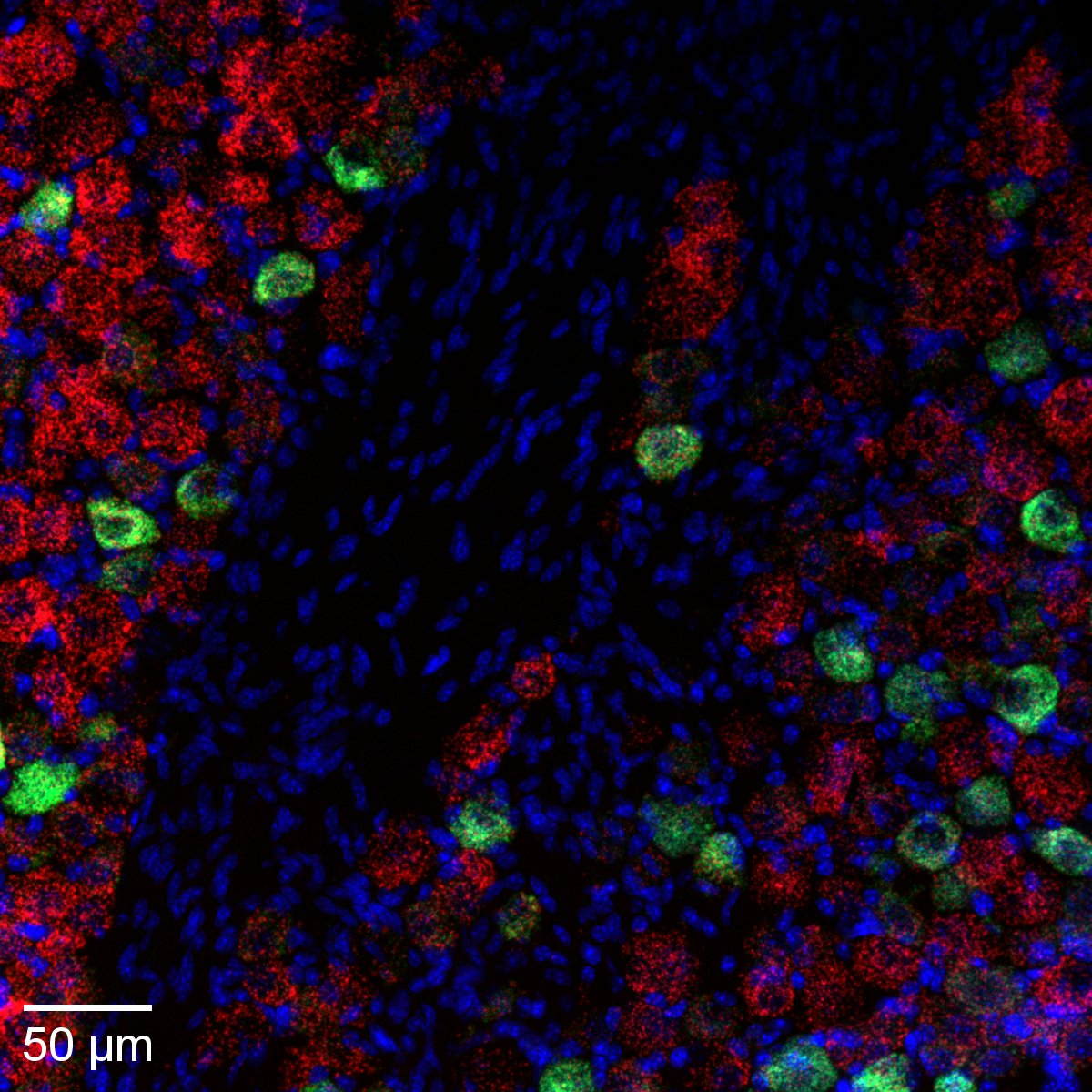
Murine femur section
Femur of a young adult mouse injected with EdU. Frozen section stained using Click-iT EdU 488 and anti-aggrecan antibody (CF555). Imaged on Nikon CSU-W1 spinning disk confocal microscope; 20x/0.75 NA. Credit: Jingzhu Zhang.
Femur of a young adult mouse injected with EdU. Frozen section stained using Click-iT EdU 488 and anti-aggrecan antibody (CF555). Imaged on Nikon CSU-W1 spinning disk confocal microscope; 20x/0.75 NA. Credit: Jingzhu Zhang.