Resources & Services
Using a modular approach, the ICO Concierge Service strives to provide streamlined access to the data and informatics infrastructure UT Southwestern research teams require to rapidly employ real-world, clinically generated data to translate discovery into better health.
The ICO’s services for researchers, learners, and faculty include:
Informatics Consultation
- Epic tools supporting translational research needs
- Clinical Decision Support Tools
- Epic registry
- Networking within CTSA resources
- Participant recruitment tools and strategies via HER
- Data set design, preparation, and implementation in Epic
Bioinformatics Support
- Next generation sequencing (NGS) data analysis: RNA-seq, ChIP-seq, scRNA-seq, etc.
- Omics data analysis: (LC-MS & LC-MS/MS) metabolomics and proteomics, (SomaScan) Proteomics
- Machine Learning: Data pre-processing, unsupervised clustering, dimensionality-reduction, feature selection, predictive models, Deep-Learning models (AI), etc.
- Medical image analysis: MRI, CT, PET, OCT, OCTA, etc.
Aamirah.Vadsariya@UTSouthwestern.edu us if you don't see the Informatics Services you are looking for.
The ICO Collaborative Process
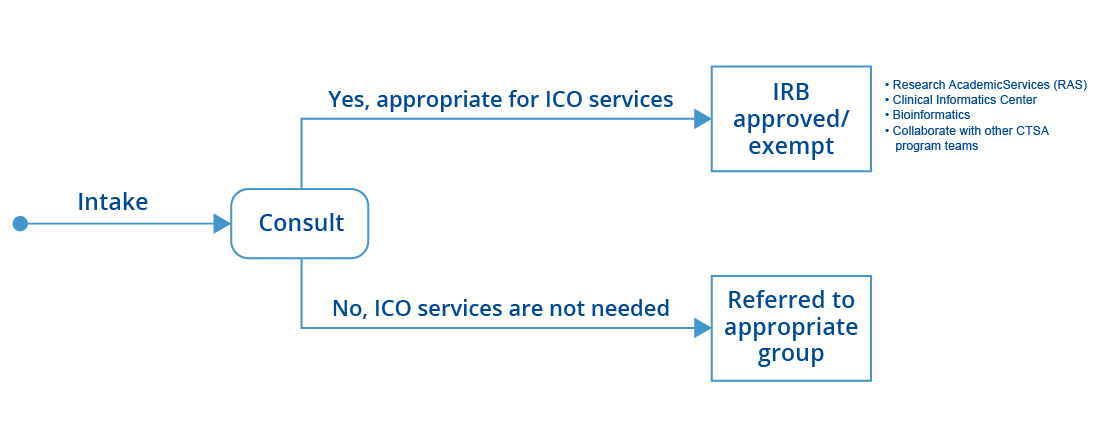
Intake
Clinicians, researchers, students, and staff with informatics needs must first schedule an ICO intake call via email. ICO consultations are by appointment only.
Consultation
Upon receipt of a consultation request, the ICO reviews the client’s needs to determine if a consultation is appropriate. That determination results in either the client’s request being submitted to our internal review board (IRB) for further action, or the client being referred to the appropriate department.
If an ICO consultation is appropriate, we will schedule a meeting with an informatician/bioinformatician to define the best options to develop for the clinical and translational research project. The consultation will include an estimate of the effort needed for the project and will clearly outline requests to make them actionable.
The ICO may link the investigator with the Research Academic Services (RAS) teams, if applicable, for clinical and translational research projects that include data extractions. If the consultation involves a request for a new Epic clinical decision support (CDS) tool or registry, we will work with our Epic team at the Clinical Informatics Center (CIC). If the researcher needs guidance on next-generation sequencing, microarrays, and functional evaluation of high-throughput data, we have a team of experienced bioinformaticians to help.
Self-Service Data Sources
- Epic SlicerDicer
SlicerDicer is a self-service reporting tool that allows researchers access to de-identified EHR data for all patients in Epic. Epic users can access SlicerDicer via the Epic Reports Menu or via the search field in the upper right corner of the interface to:
- Get real-time patient counts and information about current UTSW patients.
- See demographics and create multiple queries within one session.
- Search specific clinics to assess study feasibility.
(Request Epic SlicerDicer access via kathleen.esselink@utsouthwestern.edu.)
- Informatics for Integrating Biology and the Bedside (i2b2)
The i2b2 Clinical Research Data Warehouse (CRDW) is a data repository for health information on the current UT Southwestern patient population. Access for UTSW faculty and research staff allows researchers to:
- Search the CRDW at UTSW.
- Obtain patient counts based on criteria selected (ICD-9 or ICD-10, demographics, labs, medications, etc.).
- See demographic breakdowns.
- Refine a patient set and then request de-identified or identified data (with IRB approval) from the CRDW.
(Request i2b2 access via kathleen.esselink@utsouthwestern.edu.)
- TriNetX: The Global Health Research Network
TriNetX is a clinical data network that also provides real-world data. Researchers can perform system data queries on UTSW data or on data from the national TriNetX database. Highlights include:
- Free access to local and national data
- The ability to ingest data (PHI, LDS, de-identified) from any data source (i2b2, Epic, OMOP, etc.)
- A network that is constantly refreshed and growing
(Request TriNetX access via kathleen.esselink@utsouthwestern.edu.)
Observational Medical Outcomes Partnership (OMOP)
500+ Million Patients
OMOP is a Common Data Model (CDM) widely adopted by medical institutions across the U.S. and around the world.

3,000 Data Quality Tests
OMOP is the most curated data standard using the Data Quality Dashboard.

Observational Data at Scale
OMOP enables the data transformation to be easily distributed and massively scaled.
In 2022, UT Southwestern embarked on the Observational Medical Outcomes Partnership (OMOP) transformation process and joined the Observational Health Data Sciences and Informatics (OHDSI) network. OHDSI is an international community using empirical methods to conduct observational and reproducible research. Once this transformation is complete, access to the OHDSI network will become available through the ICO.
The goal is to transition from the Epic Caboodle dimensional model to the OMOP CDM. We will leverage publicly available transformations of the OMOP CDM to the PCORnet and Fast Healthcare Interoperability Resources (FHIR) CDMs for future development. Thus, a query formulated once can run against each CTSA Program hub’s Epic instance, with resulting datasets combined across hubs or across members of the CTSA Program network.
We will use synergistic strategies, such as collaborating with CTSA Program hub partners, to help map OMOP CDM vocabulary to our ETL framework document, preventing the need to replicate work. This will improve analytical interoperability across our CTSA Program hubs, streamlining access to integrated cross-institutional clinical data.
Cite Us
Have you received a consultation from the ICO? Remember to cite CTSA services in your publications and posters.
Suggested Citations
| Scenario | Suggested Citation |
|---|---|
| "I used the UT Southwestern CTSA Program facilities or services (e.g., UTSW CTSA Program consulting services, Informatics Coordinating Office)" OR “I received training or funding through the UT Southwestern CTSA Program (e.g., Pilot Grant Awards Program, master’s degree certificate program, etc.)" |
“Research reported in this publication was supported by the National Center for Advancing Translational Sciences of the National Institutes of Health under award Number UL1 TR003163. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health” |
For more information, contact:
Aamirah Vadsariya
CTSA Program Informatics Core Project Manager
Aamirah.Vadsariya@UTSouthwestern.edu
