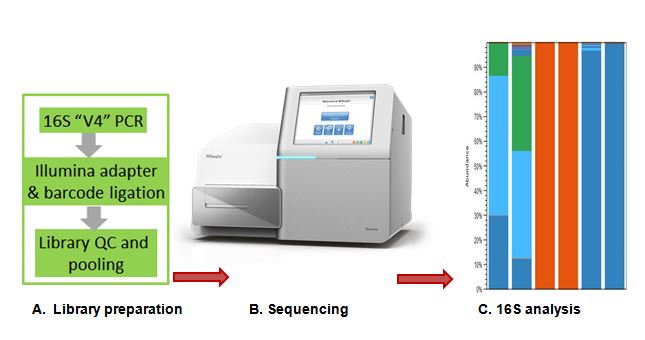
16S Sequencing
Assay Overview
We provide services to sequence various hypervariable regions (V1-V9) of 16S rRNA gene for bacterial taxonomy and phylogeny. Customer can select the hypervariable region/s based on their sample source and study aims.
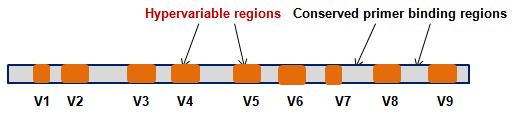
For commonly studied regions such as: V3 & V4, we use Illumina Nextera recommended primer sets and protocol to amplify the targeted region/s and ligate illumina adapter & barcodes. We also offer in house developed protocol for full length 16S rRNA gene sequencing.
16S Amplicon PCR Forward Primer = 5'
TCGTCGGCAGCGTCAGATGTGTATAAGAGACAGCCTACGGGNGGCWGCAG
16S Amplicon PCR Reverse Primer = 5'
GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAGGACTACHVGGGTATCTAATCC
Customers need to provide either original sample (to isolate DNA) or extracted genomic DNA representative of their sample. We recommend 100-500ng (20ng/ul conc.) high complexity DNA to generate high quality 16s sequencing libraries. We use illumina recommended dual indexing to barcode sequencing libraries before pooling. QC pass libraries are precisely quantified using Q PCR before loading onto MiSeqDX illumina platform using 300 PE protocol.