New tool sheds light on where RNA is found in cells
PHOTON, developed by UTSW scientists, could lead to new therapies for neurodegenerative diseases
DALLAS – June 26, 2025 – A new tool called PHOTON, developed by scientists at UT Southwestern Medical Center, can identify RNA molecules at their native locations within cells – providing valuable clues to where different RNA species are distributed spatially in response to various cellular cues. This approach, detailed in Nature Communications, could help researchers explain processes that go awry in diseases and potentially identify new targets for treatments.

“Aging and many neurodegenerative diseases impose significant stress on cells, causing a subset of cellular RNA to redistribute into various subcellular compartments such as the stress granules,” said study leader Haiqi Chen, Ph.D., Assistant Professor in the Cecil H. and Ida Green Center for Reproductive Biology Sciences and of Obstetrics and Gynecology at UT Southwestern. “PHOTON allows us to detect the spatial redistribution of cellular RNA in diseases versus health, helping us understand how these diseases cause damage to cellular functions.”
Cells make all the different proteins they need through a two-step process in which an enzyme “transcribes” the genomic molecule DNA into RNA, which is then “translated” into proteins by cellular organelles called ribosomes. Because translation happens in numerous locations throughout cells to produce proteins crucial for various functions, understanding which RNA molecules are present in those locations can shed light on how cells utilize proteins and how these processes can differ in health and disease.
Although researchers have examined RNAs present in some sturdy organelles that can be readily isolated from cells, such as the nucleus or mitochondria, Dr. Chen explained, other structures in cells are more fragile, transient, or lack membranes, preventing the use of this approach. Techniques exist to spatially identify RNA species, he added, but they can be prohibitively expensive and require extensive expertise to perform.
Seeking a better way to accomplish this goal, Dr. Chen and his colleagues developed PHOTON, short for Photoselection of Transcriptome over Nanoscale. The researchers designed DNA-based molecular cages that readily bound to all the RNA in the cells. These molecular cages can open when exposed to light, allowing for further chemical labeling.
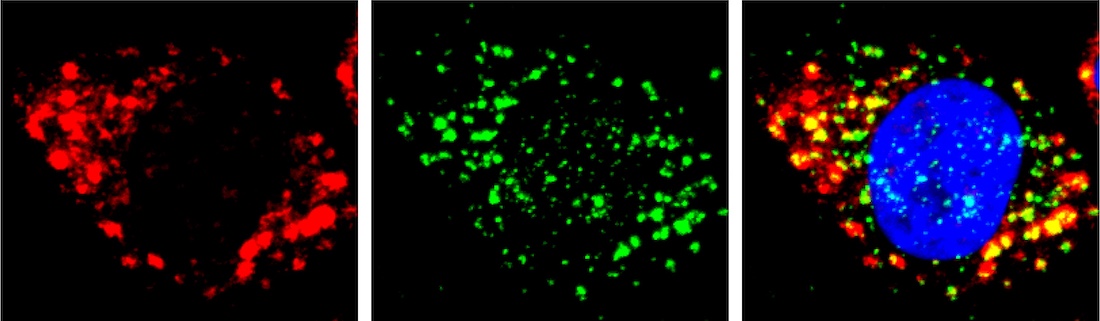
After they allowed cells under a microscope to take up these cages, the team shined an extremely narrow laser beam – only 200 to 300 nanometers across – on structures of interest, such as particular organelles. This light triggered the molecular cages to open, allowing RNAs located only in the illuminated region to be specifically labeled. Afterward, the researchers collected the labeled RNA molecules and sequenced them to learn their identities and functions.
As proof of principle, the researchers used PHOTON to examine RNAs present in the nucleolus and mitochondria, showing that the RNAs identified through PHOTON closely matched those in published databases produced by isolating these organelles from cells. Next, Dr. Chen and his colleagues applied PHOTON to stress granules – transient and membraneless structures that cells form under stress. Although most stress granule RNAs identified by the scientists matched those in published databases, they found some discrepancies using PHOTON, suggesting potential contamination in previous research.
The researchers then used PHOTON to answer a long-standing question in biology: whether a chemical modification found on some RNA molecules, called m6A, might help pull these RNAs into stress granules. By analyzing RNAs identified through PHOTON, the researchers found that those in the stress granules carried significantly more m6A than those outside them – suggesting this modification does play a role in moving specific RNAs into stress granules, which could prevent them from being translated into proteins.
Dr. Chen said he and his colleagues in the Chen Lab plan to continue using PHOTON to study the distributions of RNA in various conditions, particularly neurodegenerative diseases and aging. By comparing distributions to those in healthy cells, he said, researchers may be able to identify new targets for therapies to treat these conditions.
Other UTSW researchers who contributed to this study are first author Shreya Rajachandran, B.S., former Research Technician in the Chen Lab who is now a medical student at the University of Texas at Austin; and Qianlan Xu, Ph.D., Qiqi Cao, Ph.D., and Xin Zhang, Ph.D., Postdoctoral Researchers.
This study was funded by the Cecil H. and Ida Green Center for Reproductive Biology Sciences Endowment and a grant from the National Institutes of Health National Human Genome Research Institute (R01HG013358).
About UT Southwestern Medical Center
UT Southwestern, one of the nation’s premier academic medical centers, integrates pioneering biomedical research with exceptional clinical care and education. The institution’s faculty members have received six Nobel Prizes and include 25 members of the National Academy of Sciences, 23 members of the National Academy of Medicine, and 14 Howard Hughes Medical Institute Investigators. The full-time faculty of more than 3,200 is responsible for groundbreaking medical advances and is committed to translating science-driven research quickly to new clinical treatments. UT Southwestern physicians provide care in more than 80 specialties to more than 140,000 hospitalized patients, more than 360,000 emergency room cases, and oversee nearly 5.1 million outpatient visits a year.
