Secondary Faculty

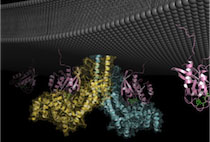
The Stone Chen Lab investigates the molecular mechanisms linking membrane signaling to cytoskeletal dynamics and their roles in human disease. Our current research centers on protein assemblies pivotal to actin polymerization and endosomal trafficking, such as the WAVE and WASH complexes, as well as the CCC-Retriever and Retromer complexes. These systems coordinate essential cellular processes such as migration, adhesion, metabolism, immune activation, and neural development.

yuhmin.chook@utsouthwestern.edu
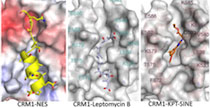
The Chook Lab studies physical and cellular mechanisms of nuclear-cytoplasmic transport. We seek to understand molecular recognition in this system and to discover new classes of nuclear localization and export signals.

In our lab, we mine large-scale data for biological discoveries. Recent research projects include: Detection and Modeling of Protein-Protein Interactions on a Proteome-wide Scale, Molecular Basis of Pathogenicity and Host Defense, Interpretation of Disease-causing Genetic Variants and Somatic Mutations in Cancer, Evolutionary Genomics of Butterflies, Genetic Basis for Unique Phenotypic Traits in Animals, and Prediction of Protein Structures and its Quality Assessment.

ankit.gupta@utsouthwestern.edu
In the Diamond lab, my research interest is generating novel reagents — nanobodies, single-chain antibodies, and antibodies — that can distinguish pathogenic and non-pathogenic conformations of tau protein and can bind to pathogenic strains of tau. I am also developing early diagnostic tools to characterize various pathogenic protein aggregates.

The Henne Lab studies how cellular membranes are sculpted during processes like vesicle budding, organelle biogenesis, and the formation of inter-organelle membrane contact sites. We employ both budding yeast and mammalian cellular systems to reveal molecular mechanisms of this membrane remodeling, and our main projects use combinations of cell biology, genetics, biochemistry, and structural biology to deeply understand cellular sculpting events.


youxing.jiang@utsouthwestern.edu
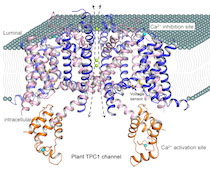
The overall research goal of the Jiang Lab is to understand the structure and function of tetrameric cation channels, utilizing a combination of membrane protein crystallography and channel electrophysiology.

lukasz.joachimiak@utsouthwestern.edu
Protein misfolding is a key element of Alzheimer’s and other neurodegenerative diseases. Our lab seeks to uncover the structure-function relationship of macromolecules involved in protein misfolding. We study how conformational changes lead proteins to self-assemble into larger structures, and how cofactors tune the formation of these structures. We believe this work will lead to breakthroughs in understanding and treating neurodegenerative diseases.

Milo's lab uses statistical mechanical and computational approaches to find performance limits of complex biological systems. These include the multi-scale mechanisms of protein folding, dynamics and aggregation (especially in neuro-toxicity and cell plasticity).
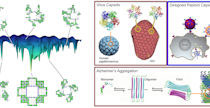

The Liu Lab studies high-order chromatin organizations and their impacts on eukaryotic transcription in both normal and diseased states. We focus on understanding the molecular and biophysical bases of the interplay between transcriptional regulation and chromatin dynamics including chromatin loop and heterochromatin formation.

Nikolaos “Nikos” Louros, Ph.D.
nikolaos.louros@utsouthwestern.edu
The Louros Lab focuses on uncovering the determining molecular forces related to protein folding, stability, and misfolding and understanding their impact on function and disease. Another facet of these important biological questions involves the process of protein self-assembly into ordered amyloid fibrils, which are the main hallmark of major neurodegenerative diseases, such as Alzheimer's and Parkinson's disease.

xuelian.luo@utsouthwestern.edu
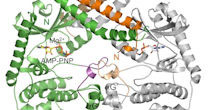
The Luo laboratory studies the molecular mechanisms of intracellular signal transduction pathways using a combination of structural and biochemical approaches. Our research program currently focuses on two major areas: the spindle checkpoint and the Hippo tumor suppressor pathway.

James.Moresco@utsouthwestern.edu
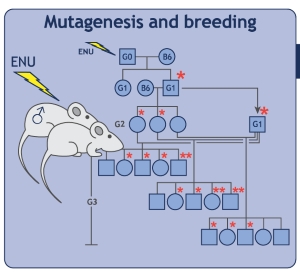
Human diseases are the focus within each of the three major research areas: the biology of the immune system, metabolism, and neurobehavioral function. The mouse phenotypic screens employed in the Center for the Genetics of Host Defense identify genes that support correct functioning of the immune system, metabolism, and the nervous system. These genes (which may be identical to the human version), or similar human relatives, protect humans from a variety of diseases including immune deficiency, autoimmunity, inflammatory bowel disease, and diabetes. In this section, the diseases, and phenotypic screens used to study them, are explained briefly. Also described are select mutations that cause or modulate one of the diseases under surveillance; the affected genes are necessary to prevent disease.

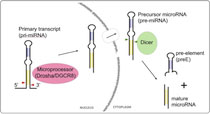
Our lab is interested in the biochemical and structural mechanisms in RNA-mediated gene regulation pathways important in development and cancer. We are currently focused on investigating how microRNAs are processed and regulated.

Binh.Nguyen@utsouthwestern.edu
In the Saelices lab, I’m using cryoEM to explore fibril structure and dreaming about solving the mystery of amyloids. We use many techniques to study the structure and mechanism of amyloid folding and deposition in disease. By determining the structure of amyloid from patient tissues, we can learn about disease-specific mechanisms that cause disease.

kimberly.reynolds@utsouthwestern.edu
The Reynolds Lab uses statistical analysis, comparative genomics, and epistasis experiments to understand the architecture and evolution of cellular systems. We are particularly interested in the evolutionary constraints on central metabolism, and large cellular structures like the flagellum.
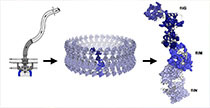

lorena.saelicesgomez@utsouthwestern.edu
We use many techniques to study the structure and mechanism of amyloid folding and deposition in disease. One of the primary methods we use to study amyloid structures is cryo-electron microscopy (cryo-EM). By determining the structure of amyloid from patient tissues, we can learn about disease-specific mechanisms that cause disease. We use additional biophysical and biochemical techniques to study the propagation, or “seeding” of new amyloid fibrils, as well as the roles short sequences play in aggregation. Altogether, we explore the process of amyloid fibril formation and how they cause disease to better understand how to fight disease in patients.


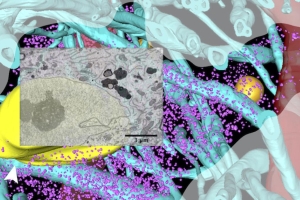
sarah.shahmoradian@utsouthwestern.edu
We study how cells shift from healthy function to early disease by directly visualizing molecular and nanoscale changes inside cells in their native environment. Using cryo-electron tomography of intact neurons and disease-relevant cell models, together with quantitative biophysics and multi-modal imaging, we examine how normally functional proteins—such as alpha-synuclein—misassemble, alter cellular compartments, and destabilize the systems that maintain neuronal health.

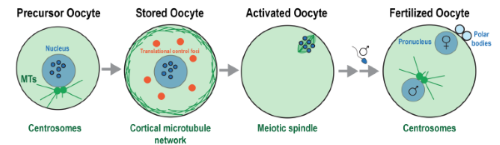
jeffery-woodruff@utsouthwestern.edu
Oocytes are life's time capsules. They can hibernate for days to decades before they experience a dramatic awakening, called activation. Once fertilized, these oocytes reprogram themselves to become a rapidly dividing embryo. Our mission is to understand the design principles of cytoplasmic reorganization during oocyte storage, activation, and fertilization.

xeuwu.zhang@utsouthwestern.edu
We study how signals are transduced across the cell membrane by using crystallography in combination with other approaches, focusing on the axon guidance receptor plexin.