Faculty and Research

Xiaochen.Bai@UTSouthwestern.edu
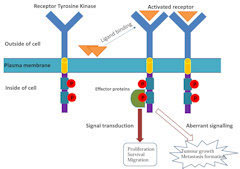
Xiaochen’s lab uses electron cryo-microscopy to study the structure of membrane protein complex in signaling transduction pathway, especially receptor tyrosine kinases which are important drug targets for cancer therapy.

chad.brautigam@utsouthwestern.edu
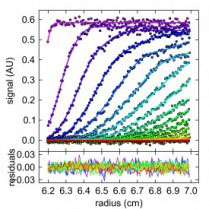
Chad’s lab uses biophysical methods to study protein-protein and protein-ligand interactions. He is also working to improve the presentation and analysis of biophysical data.

Konstantin Doubrovinski, Ph.D.
konstantin.doubrovinski@utsouthwestern.edu
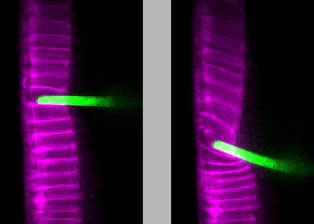
Our lab develops novel techniques to measure the mechanical properties of epithelial tissues in vivo. We combine this with genetic, imaging, and computational approaches to examine how specific cytoskeletal components interact to give rise to tissue mechanics, and how mechanical properties in turn give rise to tissue shape.

jan.erzberger@utsouthwestern.edu
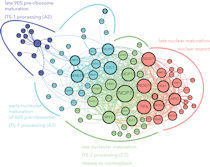
Jan’s lab is interested in understanding the dynamics of protein-RNA complexes during ribosome biogenesis. We are particularly focused on the roles of ATPases in coordinating ribosomal RNA processing and remodeling events, and in signaling between the ribosome biogenesis pathway and the cell cycle machinery.

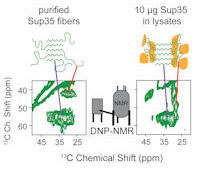
kendra.frederick@utsouthwestern.edu
Kendra's lab uses NMR spectroscopy, protein chemistry and yeast genetics to determine the structures, dynamics and energetics of protein folding in complex physiological environments such as those involved in the initiation and progression of human disease.

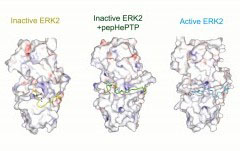
elizabeth.goldsmith@utsouthwestern.edu
Betsy's lab studies how proteins are regulated by conformational changes, focusing on protein kinases and using crystallography, NMR, mass spectrometry, and biochemistry.

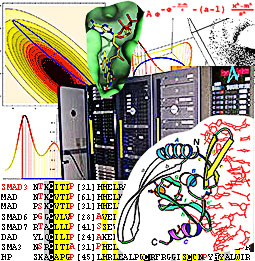
nick.grishin@utsouthwestern.edu
We work at the interface of biology, computer science, mathematics, and physics. Our group specializes in computational biology of proteins and combines sequence and structure analysis with evolutionary considerations to facilitate discoveries of biological significance.

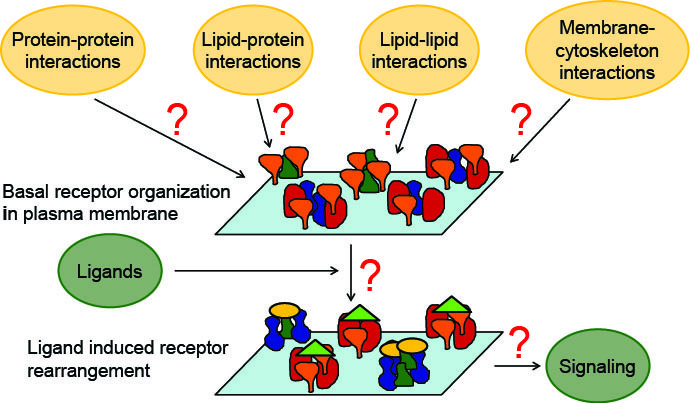
khuloud.jaqaman@utsouthwestern.edu
Khuloud's lab develops integrative approaches combining single-molecule and super-resolution imaging, computational image analysis, and mathematical modeling to investigate the contribution of cell-surface receptor organization and interactions to cell signaling.

zbyszek.otwinowski@utsouthwestern.edu
The main research focus of the Otwinowski Lab is on developing computational and statistical methods and protocols for macromolecular structure determination using X-ray crystallography.


matthew.parker@utsouthwestern.edu
Matthew’s lab utilizes a multidisciplinary approach to study the mechanisms underlying DNA replication initiation in metazoans. Insights gleaned from cellular studies guide in vitro reconstitution experiments to illuminate the molecular basis of this fundamental process

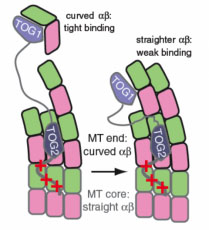
Luke’s lab studies microtubule dynamics, seeking to discover how complex behavior emerges from the biochemical properties of molecular components.

Jose’s lab studies the molecular mechanisms of intracellular membrane fusion, with particular focus on neurotransmitter release and its regulation during presynaptic plasticity.


michael.rosen@utsouthwestern.edu
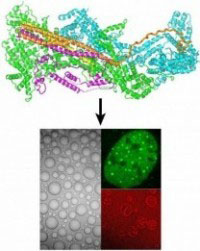
The Rosen Lab studies the physical mechanisms of actin regulation. We seek to understand both the structure and dynamics of individual pathway components and also how and why those components are organized into micron-scale cellular assemblies.

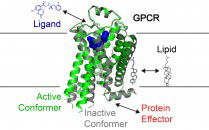
dan.rosenbaum@utsouthwestern.edu
Dan's lab studies the structure and function of eukaryotic integral membrane proteins. The goal of his research is to understand how ligands and the membrane environment influence the conformational landscape of key membrane proteins involved in cellular signaling.

daniel.stoddard@utsouthwestern.edu
Daniel uses technology of atomic-resolution single particle cryo-electron microcopy (cryo-EM) and cellular cryo-electron tomography to tackle unanswered questions in structural cell biology and to provide a new understanding of the molecular causes of diseases.

diana.tomchick@utsouthwestern.edu
Diana's lab is focused on the relationship of protein structure to biological function, primarily characterized via the technique of protein crystallography. She is also interested in improving protein crystallization techniques, crystallographic data acquisition, and experimental methods of phasing.

weiwei.wang@utsouthwestern.edu
Weiwei’s lab focuses on how ion channels associate with scaffolding proteins to form large signaling assemblies using electrophysiology, single molecule fluorescence microscopy and cyro-electron microscopy methods.
