President’s Lecture Series: ‘Zombie’ enzymes reveal mechanism of COVID-19 viral disguise

The Tagliabracci lab holds a special ritual when someone gets a paper published in a journal or wins an award. Honorees get a bottle of champagne and pop the cork to create a small dent in the ceiling’s tiles. Using a permanent marker, they circle the dimple and sign their names. That means newly arrived lab members need only look up for inspiration.
Everyone in the UT Southwestern community is invited to hear about the research behind some of the most recent dents and signatures at the next President’s Lecture. Vincent Tagliabracci, Ph.D., Associate Professor of Molecular Biology, will describe how his laboratory found a significant vulnerability in the COVID-19 virus that points toward a potential new treatment target. The talk, titled “COVID’s Camouflage,” will be at 4 p.m. on Jan. 26 in the Tom and Lula Gooch Auditorium. A reception will follow.
Known across campus as “Vinnie,” Dr. Tagliabracci, a Cancer Prevention and Research Institute of Texas Scholar, one of the University’s newest Howard Hughes Medical Institute (HHMI) Investigators, and a member of the Harold C. Simmons Comprehensive Cancer Center, would rather discuss his trainees’ successes than his own. He agreed that his ceiling is quickly filling with dents and signatures.

Among recent lab VIPs are Gina Park, an M.D./Ph.D. candidate in the University’s Perot Family Scholars Medical Scientist Training Program, and Adam Osinski, Ph.D., a postdoctoral researcher in the Tagliabracci lab. The two were co-lead authors on a study in Nature that revealed a long-elusive mechanism that coronaviruses, including SARS-CoV-2 – the one that causes COVID-19 – use to evade detection by the cells they infect.
Both HHMI and Forbes have featured this UT Southwestern work in stories, indicating its weight as a significant research advance against COVID-19.

Like many of those advances, the research was based on endeavors that predate the pandemic. The Tagliabracci lab has long studied so-called “zombie enzymes,” or pseudokinases, proteins that resemble classical kinases but appeared nonfunctional.
Turns out the zombie enzymes are only dead if researchers are looking for phosphorylation, the moving of phosphate from one molecule to another in the chemistry for which classical kinases are known.
Pseudokinases carry out a range of chemical reactions so unexpected they needed new names, such as AMPylation and glutamylation. Each of those reactions is now known to be important to cell signaling and to the spread of potentially deadly bacteria like the type that causes Legionnaires’ disease.
“No one would detect phosphorylation in them because they do something else – a different kind of chemistry,” Dr. Tagliabracci said. In the case of SARS-CoV-2, the reaction is RNAylation, he added.
Viruses cannot reproduce on their own. To survive, the SARS-CoV-2 virus must find a way to sneak into a host cell and hijack its cellular machinery to make copies of its genetic material. For the COVID-19 virus, that’s a single strand of RNA. To take over the host cell’s work, SARS-CoV-2 creates a disguise in the form of a chemical cap on its RNA that is identical to that of the host.
“People have been working on coronaviruses for 40 years, and they’ve known that the RNA has this camouflage at one end, but nobody knew how the virus created that disguise,” Dr. Tagliabracci explained. “We figured out that mechanism.”
They started working on the project when the pandemic hit.
“We realized we could try to help out somehow, especially because the coronavirus genome encoded one of these pseudokinases, known as the NiRAN domain,” Dr. Tagliabracci said. “That was what Gina and Adam worked on throughout the pandemic.”
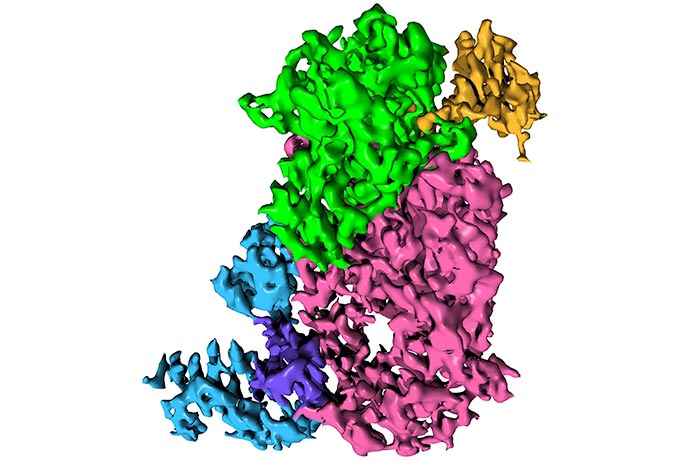
They characterized the mechanism whereby NiRAN moves the viral RNA into position for capping using another viral protein called nsp9 as an intermediate. This then allows for the final step, which ultimately forms the chemical camouflage and protects the viral genome from the host. Inhibiting NiRAN’s action brought viral replication to a halt, meaning that the pseudokinase was essential for successful infection.
If scientists could find a small molecule to inhibit the capping mechanism without affecting the host cell, it could be a novel addition to the current arsenal of antiviral drugs, Dr. Tagliabracci said. Because the virus mutates with such frequency, he added, the ideal strategy would likely be a cocktail of antivirals that attack the virus from multiple directions.
The search is on for an agent that could inhibit the RNA capping pathway that the team discovered, essentially unmasking the viral RNA so that the host cell can recognize and fight the virus. In addition, SARS-CoV-2’s unconventional capping mechanism is likely to be conserved in other coronaviruses, such as the ones that cause the common cold in humans and those that cause illness in other animals.
The lab is currently screening for drug candidates.
“We’ve got some promising hits, so I don’t think it’s too farfetched to say that in the next decade we’ll be taking a drug that targets the NiRAN domain to stop coronavirus (before it can replicate). It might not be us that comes up with it, but we’ll give it our best shot,” he said.
Dr. Tagliabracci is a Michael L. Rosenberg Scholar in Medical Research.

