Back from the dead
UT Southwestern molecular biologist conducts high-risk, high-reward research on pseudoenzymes.
I shall be telling this with a sigh
Somewhere ages and ages hence:
Two roads diverged in a wood, and I –
I took the one less traveled by,
And that has made all the difference.

Dr. Vincent Tagliabracci studies so-called “zombie enzymes” in work that is bringing those pseudokinases back from the scientific dead.
Known around campus as “Vinnie,” Dr. Tagliabracci is an Assistant Professor of Molecular Biology and a Cancer Prevention and Research Institute of Texas (CPRIT) Scholar. His interest in pseudokinases was based on logic and curiosity. If the pseudoenzymes were nonfunctional, he reasoned, why were they conserved throughout evolution?
In humans, there are more than 500 protein kinases. Classical kinases are enzymes that transfer phosphates from a high-energy molecule such as adenosine triphosphate (ATP) to proteins to regulate a multitude of cellular processes. However, about 10 percent of family members lack key amino acid residues needed for phosphorylation, thus rendering them useless for transferring phosphate groups.
But in studies published in Cell in 2018 and Science last year, the Tagliabracci laboratory found that some pseudokinases are only dead when scientists are specifically looking for phosphorylation.

“No one would detect phosphorylation in them because they do something else – a different kind of chemistry. We found that they catalyze an entirely new reaction for a member of the protein kinase superfamily,” he says. He’s referring to the Cell study in which his lab identified one pseudokinase that does AMPylation, which involves moving an adenosine monophosphate (AMP) molecule onto a protein.
His lab also reported that another pseudokinase carries out glutamylation, the transfer of the amino acid glutamate to a protein. Both pseudokinases are now considered important to cell signaling as well as to the spread of deadly bacteria.
In 2009, the laboratory of Dr. Kim Orth, Professor of Molecular Biology and Biochemistry, coined the term AMPylation and was the first to report its association with a bacterial virulence factor.
The glutamylation paper published in Science last year by Dr. Tagliabracci and an M.D./Ph.D. candidate in his laboratory revealed a major mechanism that the bacteria for Legionnaires’ disease – Legionella pneumophila – use to cause frequently fatal pneumonia.
“We found that glutamylation plays an important role during Legionella infection. It inhibits the toxic effect the bacteria usually exert on the host cell,” says lead author Miles Black, who is in the Medical Science Training Program at UTSW. “This activity seems counterintuitive at first, but it’s beneficial for Legionella to keep the host cell alive and healthy as long as possible so the bacterium has more time to replicate and extract nutrients.”
Targeting zombie enzymes
Dr. Tagliabracci’s zombie hunt begins with a bioinformatics-based search for protein kinases that seem to lack the key catalytic sequences that otherwise fit the family profile.
“These are usually so different that most people haven’t noticed that they are kinases. In fact, the amino acid sequences have diverged so much that they are not recognized by standard bioinformatic methods and they don’t show up in databases of kinases,” he says.
For that step, Dr. Tagliabracci works with Dr. Krzysztof Pawlowski, a bioinformatician from Poland and visiting Associate Professor in the Tagliabracci laboratory who specializes in searching genomes. So far, they’ve searched the human genome and those of Legionella and a plant bacterium called Pseudomonas syringae that plagues agriculture.

Dr. Tagliabracci’s career journey – and the winding path on which he happily mentors others – carries the excitement of following the science where it leads. He gravitates toward riskier projects but is aware these investigations are harder to get funded.
Luckily, the National Institutes of Health (NIH) has a grant program tailored to researchers like him. Dr. Tagliabracci is UT Southwestern’s newest winner in the NIH Director’s High-Risk, High-Reward Research Program. The NIH funds the awards for extraordinarily creative scientists to spark scientific discovery by supporting research proposals with transformative potential that might struggle to get traditional funding due to their inherent risk.
“It’s a project to study human pseudokinases to see if they aren’t really dead,” he explains of his New Innovator Award investigation called “Breathing Life into Dead Enzymes.”
Applying logic to risky science
A native of Canada, Dr. Tagliabracci did his undergraduate work in chemistry and biology at the University of Indianapolis, followed by earning a Ph.D. in biochemistry and molecular biology at Indiana University School of Medicine. For his postdoctoral fellowship, he joined the laboratory of Howard Hughes Medical Institute Investigator (HHMI) Dr. Jack Dixon at the University of California, San Diego, where he searched successfully for a casein kinase that had eluded the field for decades.
What he did, though unusual at the time, has become a hallmark of his work – using recent advances in bioinformatics to identify novel kinases.
Secreted proteins – such as the casein proteins in breast milk that provide a rich source of nutrients for newborns – have been known to be phosphorylated since 1883, but the kinase responsible for phosphorylating them was unknown. Dr. Tagliabracci is lead author of a 2012 article in Science that identified a family of secreted kinases and found that one specifically, Fam20C, is the enzyme that phosphorylates milk casein.
What he did, though unusual at the time, has become a hallmark of his work – using recent advances in bioinformatics to identify novel kinases.
“We said, ‘Let’s not restrict ourselves to the kinome (the atlas of known kinases). Let’s look at other sequences that are different enough not to be in the kinome.’ At first, we didn’t know if the ones we identified would be kinases, but we tested them and Fam20C had kinase activity. We had discovered the physiological casein kinase,” he says.
As his postdoc fellowship was coming to an end, he came to UT Southwestern to deliver a job talk, attended by Dr. Anju Sreelatha, then a graduate student in Dr. Orth’s laboratory. Dr. Orth is also an HHMI Investigator.
“After the seminar, I told Dr. Orth that I really liked Vinnie’s talk and thought his science was amazing,” Dr. Sreelatha recalls. “Kim said, ‘Oh, you should consider doing a postdoc with him.’ At the time, he was in San Diego, so I emailed him and interviewed with him while he was still in California. He decided to come here! I didn’t have to move!” she says with a laugh.
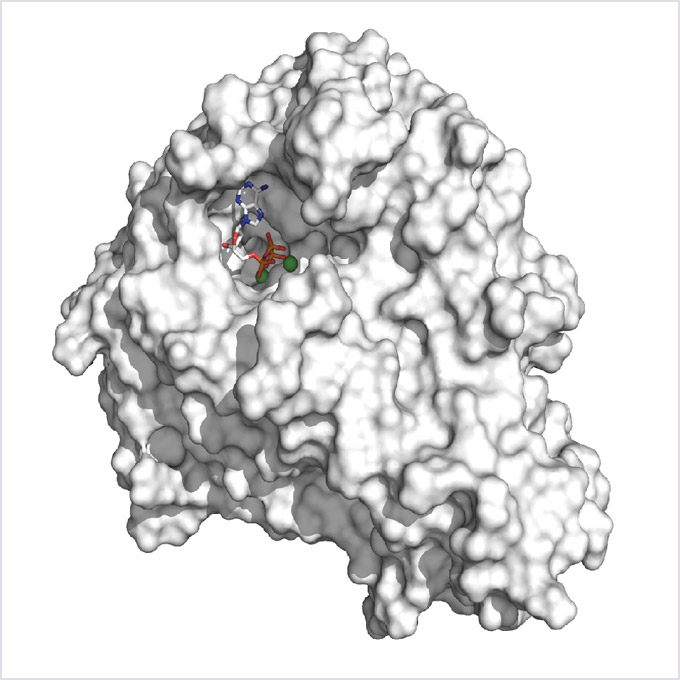
As the first postdoctoral researcher in the Tagliabracci lab, she took on the project that eventually became the Cell study described above: explaining the discovery of AMPylation in the zombie kinase selenoprotein O (SelO). A recent scientific article had listed SelO as one of the top 10 most highly conserved proteins with unknown function.
Drs. Sreelatha and Tagliabracci were interested in SelO because 1) it is found in organisms from bacteria to humans, indicating its importance throughout evolution, and 2) the human version incorporates selenium, a nutritionally essential trace element. Again, that seemed to suggest importance, Dr. Sreelatha says.
They decided to solve the structure of the protein using X-ray crystallography in the Structural Biology Core – the University’s facility for this technology – led by Dr. Diana Tomchick, Professor of Biophysics and Biochemistry.
“When we got the structure, we noticed that the ATP in the active site was a mirror image of what it should be. It was flipped,” she says. When ATP is in its normal orientation, the end phosphate is being transferred, i.e., typical phosphorylation. The researchers wondered if the ATP was transferring the AMP from the other end of the molecule.
AMPylation had been seen in other proteins, but none of them has a kinase fold, she says. “This is the first instance where a kinase fold was identified as doing something other than phosphorylation.” Remember, kinases by definition were once thought only to phosphorylate.
Identifying the undead
“We were very excited. We knew it would have been kind of groundbreaking if it were true,” she says of the finding, which opened up the field to the idea of an old fold having new activity. To confirm their finding, they used mass spectrometry and did biochemical experiments to confirm which end of the ATP molecule was being transferred.
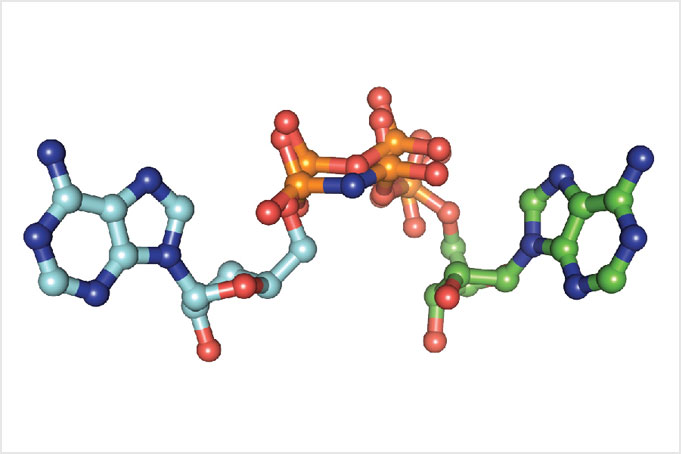
With the kinase finding, Dr. Sreelatha’s career took off. She recently set up her own laboratory and joined the faculty as an Assistant Professor of Physiology and a CPRIT Scholar.
She describes Dr. Tagliabracci as a scientific risk-taker, but a careful and logical one.
“He’s always willing to go after something that is unknown and out-of-the-box, and there is a logic behind the science. He encourages that way of thinking. He is supportive and willing to let you explore on your own,” she says.
Dr. Tagliabracci credits his advances to the tremendous power of mentoring, so he is happy to mentor others. In fact, the Molecular Biology Department has a mentoring program that pairs early career faculty with tenured researchers. He and his mentor – Dr. Orth – meet regularly to talk about their science and how to build a career.
He says those talks remind him of the importance of relationships across the University and in the lab, where the “fun” in his fundamental discoveries comes from the exhilaration of observing something no one has seen before and his attempts to keep the atmosphere upbeat.
“There are good days and bad days in the lab, just like everywhere. We have a pretty good time in the lab. It’s quite a vibrant environment, and I try to make sure everyone’s having fun. No one wants to come to work every day and be miserable. Science is not easy. It can be tough, projects fail,” he says, adding: “You learn something from every failed experiment.
“They teach you what not to do,” he says with a laugh.
Dr. Orth, an HHMI Investigator, holds the Earl A. Forsythe Chair in Biomedical Science and is a W.W. Caruth, Jr. Scholar in Biomedical Research.
Dr. Sreelatha is a W.W. Caruth, Jr. Scholar in Biomedical Research.
Dr. Tagliabracci is a Michael L. Rosenberg Scholar in Medical Research.

