Genetic work provides insight into nerve cell process called myelination
Research could have impact for future studies of multiple sclerosis, cerebral palsy, dementia, and other diseases
UT Southwestern researchers have developed a powerful genetic tool to study a process called myelination. This metabolic action involves the production of a fatty, productive layer around nerve cells to enhance their speed and efficiency in transmitting electrical signals.
Changes to the mouse DNA allowed scientists to tweak the behavior of oligodendrocytes – support cells that wrap nerve cells with a fatty insulating layer of myelin that is required for normal function. The mouse model, called Enpp6-CreER, assists basic research into the earliest stages of myelin development and enables studies of multiple sclerosis and other diseases of myelin dysfunction.
“The process of wrapping around the axon section of the nerve cell is the most mysterious stage of myelination,” said Lu Sun, Ph.D., Assistant Professor of Molecular Biology and senior author of the study, which was published recently in Nature Neuroscience. “This tool allows us to see what myelination really looks like. It opens up a way to study early stages of the process.”
With Enpp6-CreER mice, researchers can give oligodendrocytes a fluorescent glow to locate them in the brain, easily turn myelin-related genes on and off, track cell development, and distinguish between what happens during normal embryonic development, adult repair and maintenance, and disease states.
Oligodendrocytes are so numerous that they outnumber the nerve cells they support. Studying their early stages has been difficult because some research methods either affect unrelated cells or don’t provide enough detail.
The key to the new mouse model is the Enpp6 gene, which encodes an enzyme involved in myelination, among other actions. Enpp6 activity is a marker of premyelinating oligodendrocytes (preOLs), an immature stage in which the cells send out processes to “survey” nearby cells and begin wrapping nerve cells with myelin.
Dr. Sun’s team used CRISPR gene-editing technology to splice Enpp6 into existing mouse lines that allow manipulation of the inserted gene. These lines were also crossed with other mouse lines that include features such as fluorescent tags that glow when Enpp6 is active and having the ability to activate multiple genes in a single length of inserted DNA.
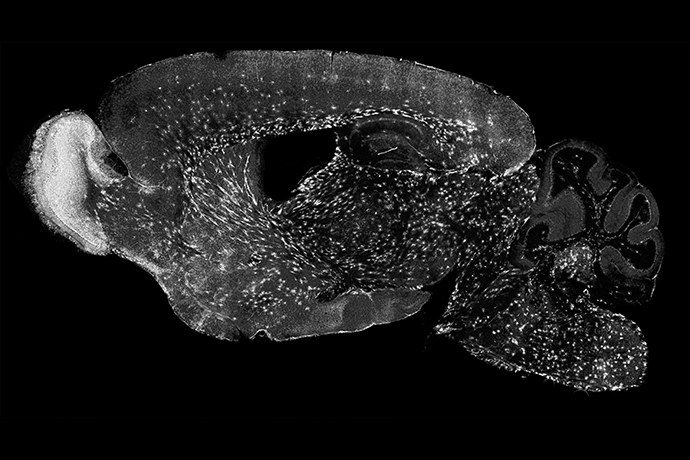
Using advanced techniques to examine gene activity in individual, genetically labeled cells from the mouse model, the researchers identified the very first step in the development of oligodendrocytes, known as committed oligodendrocyte precursor 1 (COP1). They also found that reducing nerve activity in specific brain areas – done by trimming whiskers on one side of the mouse’s muzzle to decrease sensory input – led to fewer surviving oligodendrocytes on the corresponding side of the brain. The fluorescent signals made it possible to identify single premyelinating oligodendrocytes as they became active within brain tissue.
“We answered two simple questions – where and when does myelin appear in the brain?” said Dr. Sun, who is also a member of both the Hamon Center for Regenerative Science and Medicine and the Harold C. Simmons Comprehensive Cancer Center as well as a Peter O’Donnell Jr. Brain Institute Investigator.
Using this tool, the researchers found that myelination occurs first in parts of the brain that control basic life functions such as breathing and muscle coordination. Areas like the hippocampus – involved in learning and memory – undergo myelination later in development, they found.
Finally, the mouse model allowed the research team to remove oligodendrocytes at distinct developmental states. This feature can be used to study demyelination and remyelination as a model of diseases such as multiple sclerosis.
“This genetic tool will allow refined studies of oligodendrocyte-neuron interactions,” Dr. Sun said. “In particular, this new tool will allow the detailed dissection of the interactions between neuronal activity and myelination, which often go awry in many neurological disorders, such as brain tumors and dementia.”
Other UTSW researchers who contributed to this study include first author Aksheev Bhambri, Ph.D., postdoctoral fellow; senior author Chao Xing, Ph.D., a Professor in the Eugene McDermott Center for Human Growth and Development, the Lyda Hill Department of Bioinformatics, and the Peter O’Donnell Jr. School of Public Health; Phu Thai, Research Technician; Payton Reynolds and Daniela Barbosa, graduate students; Ze Yu, Computational Biologist; and Tripti Sharma, Ph.D., a Research Scientist during the time of the study who now works at UTSW as a Commercialization Success Partner in the Office for Technology Development.
This work was supported by a UT Southwestern Endowed Scholarship, a Chan Zuckerberg Initiative Ben Barres Early Career Acceleration Award, and a Klingenstein-Simons Foundation Fellowship Award in Neuroscience.
Endowed Title:
Dr. Sun is a Southwestern Medical Foundation Scholar in Biomedical Research.

