Dissecting data to defeat disease

Dr. Yang Xie, Director of UT Southwestern’s Quantitative Biomedical Research Center (QBRC), made headlines this summer when she developed a tool to predict which patients would benefit the most from aggressive high blood pressure treatment.
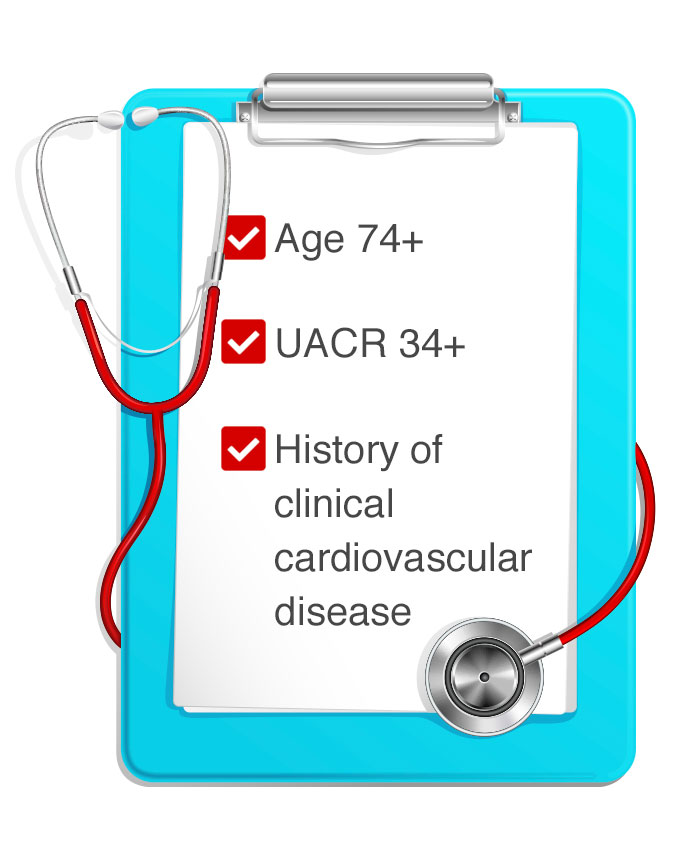
The risk prediction model, published in the American Journal of Cardiology, describes how three variables routinely collected during clinic visits – patient age, urinary albumin/creatinine ratio (UACR), and cardiovascular disease history – can identify patients for whom the benefits of intensive therapy outweigh the risks.
“Large randomized trials have provided inconsistent evidence regarding the benefit of intensive blood pressure lowering in hypertensive patients,” says Dr. Xie, corresponding author of the study and an Associate Professor of Clinical Sciences and Bioinformatics. “To the best of our knowledge, this is the first study to identify a subgroup of patients who derive a higher net benefit from intensive blood pressure treatment.”
The researchers’ algorithm determined that patients at highest risk for early, major adverse cardiovascular events were age 74 or older, with a UACR of 34 or higher, and a history of clinical cardiovascular disease. Patients meeting one or more of those criteria were among a high-risk group that had a greater benefit from intensive treatment to lower blood pressure.
In contrast, patients younger than 74, with a UACR below 34, and no history of cardiovascular disease may do as well with less intensive treatment, the researchers found.
“This model would be easy to implement in a clinical setting because it requires no additional lab tests or computational tools,” points out Dr. Xie. “We hope to design a prospective clinical trial to further validate this algorithm.”
The creation of an algorithm begins by working with clinical colleagues to identify an important question and forming a hypothesis to test. Depending on the complexity of the problems, collecting, analyzing, modeling, validating, and interpreting the data can take several months to several years, she explains.
Groundbreaking research
This wasn’t the first time that Dr. Xie, who holds both an M.D. and a Ph.D., has contributed to groundbreaking research at the intersection of data analytics and medicine. In the last few years, Dr. Xie’s team has developed several computational models to predict lung cancer patients’ outcomes based on their clinical characteristics and genomic profiles. The QBRC recently helped develop a program to estimate survival in kidney cancer patients.
“More and more, medicine is being guided by complex data analyses that only computers can do,” says Dr. Xie, who is also Director of the UTSW Bioinformatics Core Facility (BICF), which provides UT Southwestern researchers access to curated databases, software resources, bioinformatics data analysis, and training. “In my own research – which focuses on improving cancer treatments through statistical and computational analyses – using computers to work through biological and clinical data is a given.”

Dr. Xie grew up in Beijing, China, and received her medical degree from Peking University Health Science Center, a master’s degree in epidemiology from Peking Union Medical College, and a doctorate in biostatistics from the University of Minnesota-Twin Cities. She became founding Director of the QBRC in 2010.
The QBRC is part of the Department of Clinical Sciences, where it serves as a bridge between Clinical Sciences and the Lyda Hill Department of Bioinformatics, home of the newer BICF.
The BICF assists researchers who want to add innovative computational technologies to their investigations. It is a University resource staffed by a multidisciplinary team with expertise in statistics, computer science, statistical genetics and genomics, database development, and data management. The facility also serves as the primary bioinformatics, computational biology, and data analysis core for UTSW cancer research with services that include selected databases, software pipelines, educational programs, and Help Desk consultations.
In addition to UT Southwestern faculty, the BCIF has consulted with researchers at UT Dallas, Southern Methodist University, MD Anderson Cancer Center, the Cleveland Clinic, the Jackson Laboratory, and even a few pharmaceutical companies.
Next-generation sequencing
“Our most-used services are genomic data analyses and mining of public data sets, including next-generation sequencing,” says Min Kim, BICF Bioinformatics Project Manager.
“More and more, medicine is being guided by the complex data analyses only computers can do.”
Also popular: the free Help Desk. One project that started as a Help Desk request for complex genomic data mining grew into a UT Southwestern study published in Cell Reports last year, says Dr. Xie. The study identified a feedback loop that regulates a metabolite essential for many cellular processes.
Among the BICF’s projects is Kidney Cancer Explorer – an integrative database and web portal that collects clinical, genomic, pathological, and imaging data from UTSW’s Kidney Cancer Program and currently contains curated data from 2,300 patients.
Funding from the National Cancer Institute, the Cancer Prevention and Research Institute of Texas, the Lyda Hill Department of Bioinformatics, and UT Southwestern’s Harold C. Simmons Comprehensive Cancer Center help support the Facility.
On the BICF Resources page, faculty members can find a variety of free software and databases to aid in their investigations, including the Lung Cancer Explorer created by the BICF and faculty members at the QBRC. To schedule a consultation, email BICF@UTSouthwestern.edu or call the Help Desk at (214) 645-1707.