Physicians
(2013–present)

Award-winning Research
From a first-in-class kidney cancer drug to the development of new surgical and radiation approaches, a uniquely collaborative environment catalyzes transformative research.
SPORE Award...
Recognized by the National Cancer Institute with a prestigious Specialized Program of Research Excellence (SPORE) Award along with the Harvard Cancer Center.
Innovation Award...

Recognized by D CEO Magazine and Dallas Innovates for its innovation and transformational creativity as a finalist for the inaugural 2020 Healthcare Innovation Award.
Nobel Prize...

Bruce Beutler, M.D.
One of only two programs in the country developing the next generation of immunotherapies with a Nobel Prize-winning immunologist.
Expertise Improving Survival
Improving Survival Rates...
Patient survival rates three times higher for stage 4 kidney cancer and exceeding national benchmarks across all stages.
Leaders in Clinical Excellence...

Recognized with a Leaders in Clinical Excellence Program Award, the Kidney Cancer Program is setting new standards in cancer care.

Belonging
Supportive care and services helping patients navigate the cancer landscape while creating a sense of community.
Advocates...
Dedicating their time and sharing their experiences, our patient and caregiver volunteers set the tone.
Community Outreach...
Promoting educational activities to increase awareness about kidney cancer.

KCP researchers develop novel model for pediatric kidney cancer
In a study published in the Journal of Clinical Investigation, researchers at the UT Southwestern Kidney Cancer Program have unveiled a significant breakthrough in understanding and potentially treating translocation renal cell carcinoma (tRCC), an uncommon yet devastating form of kidney cancer that disproportionately impacts pediatric patients.
Translocation renal cell carcinoma, which accounts for only 5% of all renal cell carcinoma cases, assumes a more grave presence in the realm of pediatric oncology, constituting roughly one-fifth of all cases in this demographic. A rare disease in an underserved population, translocation renal cell carcinoma has long been associated with a bleak prognosis due to a lack of targeted treatment options, and it is often considered incurable in its advanced stages. Many different genes have been linked to the development of tRCC, presenting a formidable challenge in the quest for effective therapies. Furthermore, the scarcity of reliable mouse models for translocation renal cell carcinoma has hindered understanding of its biological underpinnings, leaving researchers seeking insight into the disease.
The paper, authored by Dr. Gopinath Prakasam, a postdoctoral fellow at the Brugarolas Lab within the Kidney Cancer Program, has unveiled a crucial breakthrough in understanding tRCC tumorigenesis. Dr. Prakasam’s research has revealed that genetic mutations, most associated with translocation renal cell carcinoma and driven by the enzyme Sglt2-Cre, lead to the development of highly aggressive tRCC tumors. This discovery not only provides essential insights into the potential pathways of tumor development but also lays a foundation for the development of mouse models for tRCC research.
“This finding is a significant step forward in our quest to understand and combat translocation renal cell carcinoma,” said James Brugarolas, M.D., Ph.D., director of the Kidney Cancer Program. “With this breakthrough, we have not only illuminated the elusive pathways of disease development but have also created a promising platform for future research and therapeutic advancements.”
Dr. Prakasam’s research has been supported by Joey’s Wings Foundation, as well as their partners, the Plano Pacers and Dallas DASH. The commitment to pioneering research by UT Southwestern and the support of Joey’s Wings and their partners continues to offer hope for patients affected by this devastating form of kidney cancer. As researchers delve deeper into the genetic intricacies of tRCC and develop further models to understand the disease, future studies may hold the promise of innovative treatments and renewed optimism for those facing this rare and challenging diagnosis.

Two KCP researchers receive more than $3 million in CPRIT funding
Two UTSW principal investigators received $3 million in research funding from the Cancer Prevention and Research Institute of Texas to study kidney cancer development and treatment.
Raquibul Hannan, M.D., Ph.D., Kidney Cancer Program Co-Leader of Radiation Oncology, received nearly $2 million for his proposal, Maximizing Anti-tumor Immunity through Simultaneous Activation of the Innate and Adaptive Immune System. Dr. Hannan’s proposal extends studies proposed in Project 3 of the SPORE which combines a novel drug, IMSA101, developed by Zhijian Chen, Ph.D. at UTSW, to activate the innate immune system together with radiation therapy and an immune checkpoint inhibitor in a phase 2 clinical trial. Dr. Hannan’s research seeks to explore the impact of combined activation of the innate and adaptive immune system in patients with suboptimal response to ICI treatment.


Qing Zhang, Ph.D., Associate Professor of Pathology, received over $1 million to study JMJD6-DGAT1 Signaling Axis Regulates Lipid Droplets and Tumorigenesis in ccRCC. Dr. Zhang’s research investigates the role of the JMJD6-DGAT1 pathway and lipid metabolism in ccRCC.
Additionally, UTSW researchers received over $15 million in other CPRIT awards across the board for cancer research.

KCP Patient Advocacy and Volunteer Program relaunches after COVID-19 pandemic
The Kidney Cancer Program Patient Advocacy and Volunteer Program relaunched at an inaugural meeting in the Simmons Cancer Center. Five new volunteers began serving within the Cancer Center Genitourinary Clinic after being introduced to providers and staff within the Kidney Cancer Program.
Founded in 2014 by Tony Towler and Merlinda Chelette, the Kidney Cancer Patient Advocacy Program seeks to initiate contact with newly diagnosed patients and improve their experiences while traversing a challenging diagnosis. The duo led fundraising and patient involvement initiatives within the Kidney Cancer Program, as well as offering important feedback to the program on how to improve services for patients.
Temporarily paused during the COVID-19 pandemic, the return of the Patient Advocacy and Volunteer Program aims to bring back the spirit of camaraderie and hope to those affected by kidney cancer.

Kidney Cancer Program 10th Anniversary Patient Event
The Kidney Cancer Program (KCP) recently commemorated a significant milestone, celebrating a decade of achievements in patient care and innovative research with an event for patients and community members. The event, which drew over 200 attendees, highlighted the program’s remarkable journey over the past ten years.
The event served as a platform for sharing insights gained from a decade of collaborative care and translational research. Patient and advocate testimonials, coupled with informative discussions led by esteemed faculty members, delved into a spectrum of topics surrounding kidney cancer treatment modalities and intricacies.
Dr. James Brugarolas, Director of the Kidney Cancer Program, outlined the remarkable advancements in kidney cancer research since the program’s inception with a comprehensive overview of the strides made in understanding and treating this disease.
A panel discussion, featuring both a patient and the faculty who collaborated to treat him, provided a deep dive into the complexities of managing challenging cases, offering diverse perspectives and insights crucial in the battle against kidney cancer. Members of the community could also attend breakout sessions to learn more about specific topics in kidney cancer care and research, including the genetic component of kidney cancer, radiology tools, multiple treatment methods, and caring for the caregiver.
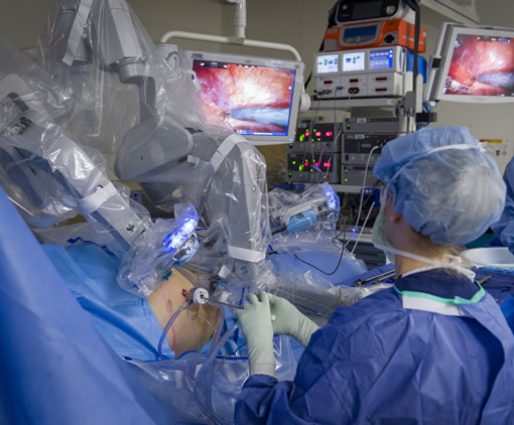
The highlight of the event was the keynote address by Dr. Vitaly Margulis, which included remarkable footage of robotic surgery, showcasing cutting-edge technological advancements in the field.
Additionally, the event received a generous boost with the presentation of a check for a $50,000 donation from local representatives of Joey’s Wings Foundation. The contribution is earmarked for pediatric kidney cancer research, emphasizing the collective commitment to advancing treatments for younger patients.
Dr. Brugarolas expressed gratitude and emphasized the importance of such events in driving progress.
“This event stands as a testament to the unwavering dedication of our team, our collaborators, and the community in the fight against kidney cancer,” he said. “The last decade has been transformative, and events like these reaffirm our commitment to continued innovation and progress.”
The Kidney Cancer Program at UT Southwestern Medical Center is one of only two NCI-designated Specialized Programs of Research Excellence (SPORE) in kidney cancer. Established in 2013, the program has become one of the largest kidney cancer efforts worldwide.

Dr. Jue Wang joins the KCP as co-Leader of Education

The Kidney Cancer Program is excited to announce the appointment of Jue Wang, M.D., as the new Co-Leader of Education. Dr. Wang succeeds Dr. Scott Schwartzman, who served in this role until recently.
With an unwavering commitment to education, Dr. Wang brings a wealth of experience and expertise to the Kidney Cancer Program (KCP). He joins the faculty of UT Southwestern and KCP as a professor of internal medicine in the division of hematology/oncology. Dr. Wang’s background spans from fundamental to clinical research and he has served as principal investigator of multiple clinical trials.
After earning his medical degree at Suzhou Medical College in China, Dr. Wang completed advanced cancer research training at both the Institute of Cancer Research of the Royal Marsden Hospital in England and the Sealy Center for Molecular Hematology and Oncology at the University of Texas Medical Branch in Galveston, where he also completed a residency in internal medicine, and fellowships in both hyperbaric medicine and medical oncology. He is the recipient of multiple accolades including a Faculty Service Award from the University of Arizona College of Medicine, a Patient’s Choice Award from the Cancer Institute at St. Joseph’s Hospital (Arizona), and a Silver Certificate of Excellence for clinical research contributions from the National Cancer Institute.
As co-Leader of Education, Dr. Wang will play a pivotal role in the KCP by implementing innovative curriculum strategies, mentoring, and supporting aspiring medical professionals, and enhancing our educational offerings.
With a deep understanding of the evolving healthcare landscape, Dr. Wang aims to enhance medical education, ensuring KCP trainees are equipped with the knowledge, skills, and compassion necessary to excel in their careers and make a positive impact on society. He is committed to fostering an inclusive learning environment that embraces diversity and encourages interdisciplinary collaboration.
“We are delighted about Dr. Wang’s appointment as Co-Leader for Education at the KCP,” said James Brugarolas, M.D., Ph.D., director of the Kidney Cancer Program. “The breadth of experience and commitment he offers to our trainees make him a great addition to the KCP leadership. I’m also grateful to Dr. Scott Schwartzman for serving in this role, and for the assistance he has provided to many trainees seeking to enhance their research skills despite his many obligations.”
Please join us in extending a warm welcome to Dr. Wang and a sincere thank you to Dr. Schwartzman.
The Kidney Cancer Program at UT Southwestern Medical Center is one of only two NCI-designated Specialized Programs of Research Excellence (SPORE) in kidney cancer. Established in 2013, the program has become one of the largest kidney cancer efforts worldwide.

Eleven UTSW investigators receive millions in DoD awards for kidney cancer research
Eleven UT Southwestern Kidney Cancer Program investigators received nine awards from the U.S. Department of Defense Kidney Cancer Research Program (KCRP) in 2023, setting a record for the most proposals recommended for funding of any institution. A third of all proposals recommended for funding were from the KCP.

Kiyoshi Ariizumi, Ph.D., professor of dermatology and immunology, received an Idea Development Award. Dr. Ariizumi’s proposal, entitled “Tumor Extravasation in Zebrafish as a Prognostic Marker and a Therapeutic Target for Metastasis of Kidney Cancer,” intends to determine if tumor extravasation in zebrafish models can be used as a biomarker for risk of metastasis in kidney cancer.
Kenneth Chen, M.D., assistant professor of pediatrics, received an Idea Development Award to investigate pediatric Wilms tumors. Dr. Chen’s project, “DROSHA regulates mesenchymal expression and chemosensitivity in Wilms tumors,” will evaluate how DROSHA regulates kidney formation as well as its role in Wilms tumors and chemotherapy response.

Ian Corbin, Ph.D., associate professor at the Advanced Imaging Research Center, received an Idea Development Award for his proposal “Leveraging SCARB1 Overexpression for the Treatment of ccRCC with Low-Density Lipoprotein Nanocarriers,” to investigate the effectiveness of low-density lipoprotein (LDL) drug delivery in animal models of kidney cancer.


Lindsay Cowell, Ph.D., associate professor at the Peter O’Donnell Jr. School of Public Health, and Payal Kapur, M.D., professor of pathology and urology, received a Translational Research Partnership Award for their proposal “Leveraging Biophysicochemical Motifs in T Cell Receptor Antigen Binding Regions and Antigen Co-occurrence to Predict Response to Immune Checkpoint Inhibitors.” This project aims to study T cells in ccRCC and determine whether they can predict the response to immune checkpoint inhibitors, one of the most common treatments for kidney cancer.

Weibo Luo, Ph.D., associate professor of pathology and pharmacology, received an Idea Development Award. “Characterization of Epigenetic and Metabolic Vulnerability in VHL-deficient ccRCC and its Therapeutic Potential,” aims to evaluate the potential of a synthetic epigenetic drug to treat clear cell renal cell carcinoma (ccRCC).

Srinivas Malladi, Ph.D., assistant professor of pathology, received an Idea Development Award. “Delineate Tumor Immune Contexture that Shapes Metastatic Progression and Response to Immunotherapy” seeks to investigate the contribution of the tumor microenvironment in immunotherapeutic response in kidney cancer.


Xiankai Sun, Ph.D., Director of the Cyclotron & Radiochemistry program, and James Brugarolas, M.D., Ph.D, Director of the Kidney Cancer Program, received a Translational Research Partnership Award to investigate PET imaging of HIF-2α in renal cancer. Drs. Sun and Brugarolas plan to leverage their understanding of HIF-2α to develop a second-generation radiotracer to visualize its expression in ccRCC patients.

Andrew Wang, M.D., professor of radiation oncology received an Idea Development Reward for his proposal “Multifunctional Immunotherapy Particle to Enable Innate Immunotherapy for Kidney Cancer,” which will investigate the use of multifunctional immunotherapy particles (MINPs) to activate natural killer cells that inhibit ccRCC tumors and enhance existing immunotherapy protocols.

Chen Yao, Ph.D., assistant professor of immunology, received the Academy of Kidney Cancer Investigators – Early Career Scholar Award to investigate stem cell-like T cells as a target for immunotherapy in kidney cancer. Mentored by Drs. Celeste Simon (University of Pennsylvania) and Brugarolas, Dr. Yao’s proposal aims to explore the role of stem-like immune cells against kidney cancer.
The Kidney Cancer Program within the Simmons Cancer Center at UT Southwestern Medical Center is one of only two NCI-designated Specialized Programs of Research Excellence (SPORE) in kidney cancer. Established in 2013, the program has become one of the largest kidney cancer efforts worldwide.

Kidney Cancer Program to celebrate 10th anniversary with faculty symposium
The Kidney Cancer Program at UT Southwestern’s Harold C. Simmons Comprehensive Cancer Center will be hosting a faculty symposium to celebrate its 10-year anniversary.
Featuring more than 60 speakers and panelists and set to include a wide range of topics from basic science to translational research and clinical trials, the symposium will offer a unique opportunity to learn about transformational discoveries, which have led to, among others, a first-in-class drug now FDA approved. It will also provide an opportunity for networking, learning about program resources, and creating new collaborations.
Topics include groundbreaking discoveries about the genetics of kidney cancer, invasion and metastases, tumor latency, metabolism, and rarer tumor types.
“This is an important milestone for the Kidney Cancer Program, and I cannot think of a better way to celebrate it than with a symposium featuring the many faculty at UTSW whose research and contributions are advancing the care of our patients,” said James Brugarolas, M.D., Ph.D., director of the Kidney Cancer Program.
The symposium, a whole-day event, will take place on Wednesday, March 29. Registration closes March 14 at noon.
The Kidney Cancer Program at UT Southwestern Medical Center is one of only two NCI-designated Specialized Programs of Research Excellence (SPORE) in kidney cancer. Established in 2013, the program has become one of the largest kidney cancer efforts worldwide.

Kidney Cancer Program receives $11 million SPORE renewal from NCI
The UT Southwestern Kidney Cancer Program Specialized Program of Research Excellence (SPORE) has been renewed by the National Cancer Institute (NCI). Initially awarded in 2016, the Kidney Cancer Program SPORE seeks to translate discoveries at UT Southwestern Medical Center into advances in the care of patients with kidney cancer.
The Kidney Cancer Program at UT Southwestern (KCP) is one of only two programs specializing in kidney cancer research nationwide, to be recognized by NCI with such an award.
Under the overall guidance of James Brugarolas, M.D., Ph.D., director of the Kidney Cancer Program, and Payal Kapur, M.D., 3 teams of investigators will delve into the most promising therapeutic areas – immunotherapy, targeted therapy, and metabolism. In addition, seed funding is available to explore new areas through the Development Research and Career Enhancement Programs. 35 pilot projects were funded during the previous SPORE cycle.
Project 1, led by Dr. Brugarolas and Xiankai Sun, Ph.D., Director of the Cyclotron and Radiochemistry program, will leverage innovative siRNA technology to develop a second-generation HIF-2α inhibitor and an innovative radiology test to monitor HIF-2α in patients. This project builds upon the research success that led to the development and subsequent FDA approval of a groundbreaking HIF-2α inhibitor, Belzutifan, during the first cycle of SPORE funding.
Drawing upon the In Vivo Metabolism Lab set up during the previous SPORE cycle, Project 2 investigators Ralph DeBerardinis, M.D., Ph.D., and Kevin Courtney, M.D., Ph.D., will evaluate new approaches to exploit glutamine addiction in renal cancer. By bypassing the limited efficacy of glutaminase inhibitors, the project aims to offer innovative solutions.
Project 3 capitalizes on the research of Zhijian Chen, Ph.D., recipient of the Breakthrough Prize. Led by Drs. Chen, Raquibul Hannan, M.D., Ph.D., and Hans Hammers, M.D., Ph.D., researchers intend to simultaneously activate the innate and adaptive immune system using a novel drug IMSA101 in combination with radiation and an immune checkpoint inhibitor. By activating both ‘arms’ of the immune system, investigators seek to maximize anti-tumor activity.
These research programs are supported by four avant-garde core facilities, encompassing pathology, imaging, data analytics, and administration.
“Discoveries by the SPORE team have been paradigm setting and practice changing,” said Carlos L. Arteaga, M.D., Director of the Simmons Comprehensive Cancer Center. “With this renewal, some of the most urgent questions in kidney cancer will be addressed. We appreciate the National Cancer Institute support of this engine of discovery, innovation, and translation to advance patient care.”

Results from Phase 2 clinical trial aim to set new paradigm of treatment for metastatic kidney cancer
A phase 2 clinical trial spearheaded by UT Southwestern Kidney Cancer Program researchers reported that stereotactic ablative radiation therapy (SAbR) effectively targets and treats oligometastatic kidney cancer while minimizing the negative impact on patients’ quality of life.
With over 400,000 new cases of kidney cancer diagnosed worldwide each year and 30% of all patients developing metastases, finding effective treatment options for oligometastatic kidney cancer is of utmost importance. Currently, there is no established standard treatment for this specific form of the disease, characterized by metastases at fewer than 5 sites. Most patients are subjected to immunotherapeutic or targeted drugs, which can cause systemic toxicity and lead to adverse effects that significantly reduce their quality of life.
Building upon pioneering efforts at UT Southwestern Kidney Cancer Program with SAbR —a precise radiation therapy technique that employs targeted, narrow beams directed at tumor sites— researchers initiated a phase 2 clinical trial. This trial, led by Raquibul Hannan, M.D., Ph.D., Chief of the Genitourinary Radiation Therapy Service, Robert Timmerman, M.D., Chair of Radiation Oncology, and James Brugarolas, M.D., Ph.D., Director of the Kidney Cancer Program, aimed to evaluate the efficacy of SAbR in controlling oligometastatic disease without the need for systemic therapy. Remarkably, 90% of the trial participants achieved metastatic disease control for over a year without any systemic therapy, and none reported serious side effects or a negative impact on their quality of life.
Building upon this and other similar results, the National Cancer Institute is funding a phase 3 clinical trial that aims to compare SAbR to medication therapy for patients with oligometastatic kidney cancer. Led by
Dr. Hannan said, if successful, the phase 3 trial will establish a standard of care for patients with oligometastatic kidney cancer for the first time.
Additional trials, also led by Dr. Hannan, aim to further establish the effectiveness of SAbR as a treatment for oligoprogressive kidney cancer and tumor thrombi that have spread to the inferior vena cava.
The Kidney Cancer Program at UT Southwestern Medical Center is one of only two NCI-designated Specialized Programs of Research Excellence (SPORE) in kidney cancer. Established in 2013, the program has become one of the largest kidney cancer efforts worldwide.

Dr. Janie Qin receives $1M Frenkel Endowed Scholar Award

Janie Qin, M.D., Assistant Professor of Internal Medicine/Hematology-Oncology and Kidney Cancer Program (KCP) member, was awarded the Eugene P. Frenkel, M.D., Endowed Scholar Award in Clinical Medicine to support the development of novel radiology tests for kidney cancer patients.
Dr. Qin joined the Simmons Comprehensive Cancer Center and KCP upon completing her fellowship in Hematology and Medical Oncology at the Icahn School of Medicine at Mount Sinai, New York, in September 2021. With the $1 million award, Dr. Qin will be driving the development of new molecular probes to characterize kidney cancer in patients.
Dr. Qin is currently co-principal investigator of two clinical trials evaluating novel radiology probes. A PD-L1 probe seeks to evaluate the expression of this most important checkpoint protein in kidney cancer patients and assess its potential in predicting response to immunotherapy (NCT04006522). A second trial focuses on the evaluation of HIF-2α in tumors (NCT04989959). HIF-2α is the target of belzutifan, an FDA approved drug for the treatment of familial kidney cancer, which was developed by Peloton Therapeutics in the UTSW BioCenter based on lead compounds discovered at UTSW. Previous research by the Brugarolas Lab suggests that HIF-2α levels in tumors may be predictive of response to drugs such as belzutifan. Both studies represent a partnership with Xiankai Sun, Ph.D., Director of the Cyclotron and Radiochemistry Program, who is synthesizing the probes.
Dr. Qin follows on the footsteps of Hans Hammers, M.D., Ph.D., Associate Professor of Internal Medicine/ Hematology-Oncology, KCP Co-Leader of Immunotherapy, and inaugural recipient of the Frenkel Scholar Award.
“Janie is a most meritorious recipient working on a cutting-edge research area and we are delighted of her selection by the Frenkel Award Committee,” said KCP Director, James Brugarolas, M.D., Ph.D. “Awards like this are instrumental in launching the careers of junior faculty and enable us to recruit investigators with the greatest promise.”

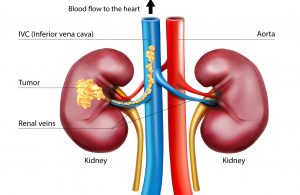
International partnership evaluates UTSW-developed approach to locally advanced kidney cancer
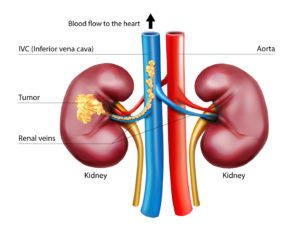
Up to 15 percent of kidney cancer patients are diagnosed with tumor thrombus, a locally advanced form of the disease where the tumor extends into the renal vein progressing sometimes into the inferior vena cava (IVC), the body’s largest vein. Once in the IVC, the tumor has open passage to the heart.
Surgery is the standard of care, but operating on such tumors is challenging, particularly if they have reached the heart. One third of the patients experience complications and mortality can be as high as 15 percent.
“These extensions can be problematic. They can block drainage of the blood into the heart causing backup, liver congestion, and swelling,” says Vitaly Margulis, M.D., a urological surgeon with expertise in this type of surgery.
In 2015 , Kidney Cancer Program (KCP) investigators developed a novel approach involving stereotactic ablative radiotherapy (SAbR). By targeting the tumor thrombus with high doses of radiation delivered from multiple angles, researchers were able to show that tumor thrombus could be controlled.
While the approach is still being evaluated in a clinical trial at UTSW (NCT02141919), an unmet medical need has led to its early adoption by institutions across the world.
A recently published report chronicles initial findings from the global experience. Fifteen patients with a tumor thrombus reaching the IVC were treated with SAbR at six leading centers in the US, UK, Italy and Australia. In 50 percent of patients the tumor thrombus approximated or invaded into the heart. About half of the patients were poor candidates for surgery and in 20 percent the tumor came back after surgery.
SAbR controlled the tumor thrombus in over 80 percent of patients relieving them from symptoms including liver congestion, swelling, bleeding and pain. While medication therapy may have contributed to thrombus control, similar benefit was observed in a subset of patients that received only SAbR.
“We are delighted to see that our pioneering efforts are benefiting patients across the world,” said Raquibul Hannan, M.D., Ph.D., co-Leader of radiation oncology at the KCP.
“This is an example of the innovation and team approach that characterizes the KCP,” said Kidney Cancer Program Director James Brugarolas, M.D., Ph.D.

DoD awards $1.3 million to UTSW investigators for kidney cancer research


Two UTSW Kidney Cancer Program investigators have been awarded more than $1.3 million in research funding from the Department of Defense Congressionally Directed Kidney Cancer Research Program.
Peter Ly, Ph.D., Assistant Professor of Pathology and Cell Biology, received an Early Career Investigator Award for $1,189,000 to explore how recurrent chromosomal abnormalities drive the development of clear cell renal cell carcinoma (ccRCC), the most common form of kidney cancer.
Gopinath Prakasam, Ph.D., a Postdoctoral fellow with the Brugarolas Lab, received a Concept Award for $123,000 to assess a novel therapeutic approach against translocation renal cell carcinoma (tRCC) a rare and highly aggressive subtype of kidney cancer largely found in children and young adults.

Pioneer in kidney cancer radiation and long-term KCP collaborator named new chair of radiation oncology

Robert D. Timmerman, M.D., has been named the new Chair of the Department of Radiation Oncology.
An expert in diseases of the central nervous system, including cancer of the brain and spine, as well as lung cancer, Dr. Timmerman has championed the use of precise, noninvasive radiosurgical tools to deliver radiation. He is credited for leading the development of stereotactic ablative radiotherapy (SAbR), also called stereotactic body radiation therapy (SBRT), which uses sophisticated image guidance and tracking to improve radiation therapy. This seminal contribution is widely regarded to have transformed the field of radiation oncology.
Dr. Timmerman has spearheaded new applications of radiation therapy for kidney cancer, most notably the first report for SAbR for inferior vena cava (IVC) tumor thrombus.
Beyond the conventional uses for bone and brain metastases in the Kidney Cancer Program (KCP), SAbR is being investigated for the treatment of small renal masses; tumor thrombi ˗ regionally advanced tumors that have grown into the IVC; oligommtastases, oligoprogression, and in combination with immunotherapies. Most recently, radiation oncology efforts have centered around PULSAR, a strategy that seeks to maximize the immunological effects of radiation therapy.
Dr. Timmerman joined UT Southwestern in 2004 as Professor and Vice Chair for Clinical Affairs in the Department of Radiation Oncology and Director of the Annette Simmons Stereotactic Treatment Center. He also served as Director of Clinical Research since 2014.
He graduated from Iowa State University with a bachelor’s degree in nuclear engineering and from the University of Tennessee with a master’s degree in reactor physics. After finishing medical school at the University of South Dakota, he completed a residency in Radiation Oncology at The Johns Hopkins Hospital.
He is a Fellow of both the American Society for Radiation Oncology and the American College of Radiology. In 2019, Dr. Timmerman received the Patricia and William L. Watson Jr., M.D. Award for Excellence in Clinical Medicine.

Higher doses of cabozantinib may offer hope to kidney cancer patients with few other options
Metastatic kidney cancer is largely incurable. Despite the growing landscape of available drugs, most tumors eventually develop resistance, with each successive line of therapy generating progressively lower returns. Some patients eventually run out of options. Investigators with the Kidney Cancer Program (KCP) at UT Southwestern’s Harold C. Simmons Comprehensive Cancer Center report that increasing the dose of an already FDA-approved medication may benefit some patients.
Cabozantinib (Cabometyx®), is one of the most potent drug therapies currently available for kidney cancer and is administered at 60 mg daily. In a manuscript published today, KCP investigators report that increasing cabozantinib by 30% controlled tumors that had progressed on the drug’s conventional dosing. Investigators found that increasing to 80 mg kept kidney cancer under control for up to 2.5 years.
“This higher dose of cabozantinib is not that different from what is used for thyroid cancer,” said Roy Elias, M.D., an author on the report who is a former KCP researcher and is currently a medical oncology fellow at Johns Hopkins Sidney Kimmel Cancer Center.
As part of the study, investigators reached out to 14 leading kidney cancer institutions, both domestic and internationally, and identified a Canadian program that had attempted a similar strategy. Tackling the same dilemma of lack of treatment options, Dr. Georg Bjarnason, M.D., medical oncologist and senior scientist at Odette Cancer Centre in Toronto, deployed the same dose-escalating approach for patients, observing similar success.
Investigators noted that the strategy is unlikely to work for patients with primary resistance and can only be attempted in patients who tolerate the standard 60 mg dose.

PATIENT SPOTLIGHT:
UTSW kidney cancer patient Kerry Gabel was one of half a dozen patients who benefited from the approach. A high school golf coach and father of two teenage daughters, Mr. Gabel was diagnosed with stage IV kidney cancer in 2016. A textbook case, Mr. Gabel experienced drug resistance on 4 therapies, including dual immunotherapy, prior to starting cabozantinib in 2018. He was on conventional doses of cabozantinib for over a year, but his disease progressed. At that point, he had progressive disease in the lung, suspected liver involvement, and one of the chest metastases appeared to invade the heart. He also developed several brain metastases. Brain metastases were treated with radiosurgery and after a discussion, and given limited availability of other treatment options, the patient elected to increase cabozantinib to 80 mg. Today, 2.5 years later, his cancer remains under control and his overall burden of disease is lower than at the outset.
“The findings suggest that this strategy could be helpful for some patients,” said Kidney Cancer Program Director James Brugarolas, M.D., Ph.D., “but clinical trials will be needed to formally evaluate the potential of the approach.”
Clinical Trial Development
KCP investigators are currently designing a trial to evaluate the effectiveness of cabozantinib 80 mg in kidney cancer patients progressing on conventional doses. The trial is anticipated to open in the last quarter of 2022 at the UTSW Simmons Comprehensive Cancer Center and at Baylor Scott & White Charles A. Sammons Cancer Center, both in Dallas.
Kidney Cancer Patient Story – Kerry Gabel
Husband, father, and high school PE teacher Kerry Gabel shares his story on battling aggressive metastatic kidney cancer and how increasing a medication beyond the conventional dosage saved his life when he was running out of treatment options.

CPRIT awards more than $2 million in research funding to UTSW kidney cancer investigators


Two UTSW Kidney Cancer Program investigators were awarded $2.2 million in academic research funding from the Cancer Prevention and Research Institute of Texas (CPIRT).
Thomas Carroll, Ph.D., received $1,040,229 to study the underlying mechanisms of sarcomatoid differentiation in renal cell carcinoma. Satwik Rajaram, Ph.D., was awarded $1,172,136 to dissect intratumor heterogeneity in kidney cancer using deep learning AI (artificial intelligence).
In total, CPRIT awarded 74 cancer research and prevention grants, of which 65 were for academic research being conducted at 10 institutions. Among the 65 investigators awarded, 16 were from UTSW, garnering more than $17 million in combined research funding.
Dr. Satwik’s award was one of only three academic research grants bestowed for computational systems biology, spotlighting UTSW’s efforts on behalf of kidney cancer in this area. PRESS RELEASE